Difference between revisions of "Phenote"
From phenoscape
| (5 intermediate revisions by 2 users not shown) | |||
| Line 3: | Line 3: | ||
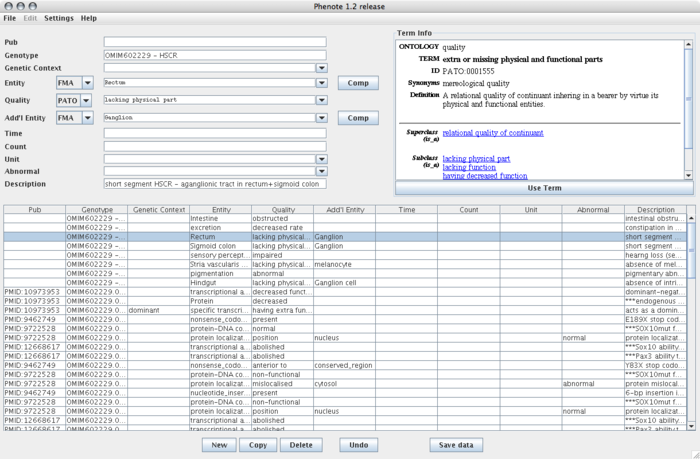
Here is sample data in the default configuration: | Here is sample data in the default configuration: | ||
| − | [[Image:Phenote-default.png|center| | + | [[Image:Phenote-default.png|center|700px]] |
| + | |||
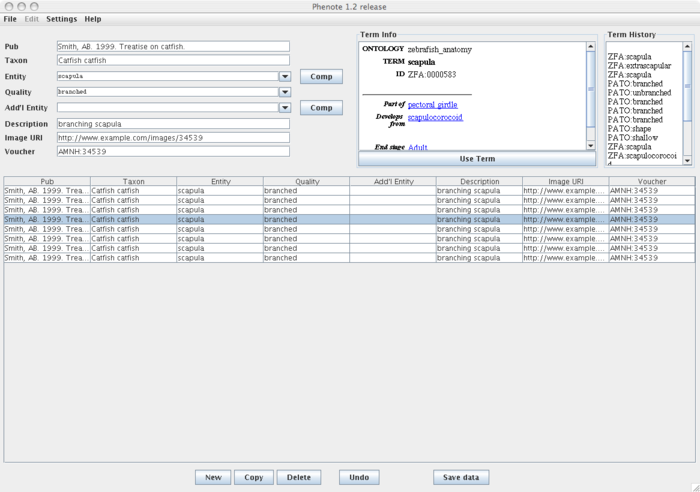
| + | The input fields in Phenote are highly configurable. Here is a sample of how Phenote might be configured for the PhenoScape project: | ||
| + | |||
| + | [[Image:Phenote-phenomap.png|center|700px]] | ||
| + | |||
| + | This configuration file can be downloaded [[Media:phenomap.cfg.zip|here]]. Phenote configurations are described in the [http://www.phenote.org/content/howtos/customize.shtml documentation]. | ||
| + | |||
| + | This interface ("list of states") may not be optimal for the PhenoScape project, especially if a set of species are expected to be coded for a given set of characters. | ||
| + | |||
| + | [[Category:Informatics]] | ||
| + | [[Category:Curation]] | ||
| + | [[Category:EQ Annotation]] | ||
Latest revision as of 23:57, 10 January 2009
Phenote is being used for phenotype annotation of mutants within model-organism projects. It allows the creation of a simple list of phenotype statements, rather than a species-by-character matrix.
Here is sample data in the default configuration:
The input fields in Phenote are highly configurable. Here is a sample of how Phenote might be configured for the PhenoScape project:
This configuration file can be downloaded here. Phenote configurations are described in the documentation.
This interface ("list of states") may not be optimal for the PhenoScape project, especially if a set of species are expected to be coded for a given set of characters.