Main Page
We will be hosting a "Needs Analysis Workshop" for this project on Sep 17-18, 2007.
Contents
Linking Evolution to Genomics Using Phenotype Ontologies
About this project
What are the developmental and genetic bases of evolutionary differences in morphology across species? Currently it is difficult to approach this question due to a lack of computational tools that allow researchers to integrate developmental genetic and comparative morphological/anatomical data.
We are addressing this by developing a database of evolutionarily variable morphological characters for a large clade of fishes (the Ostariophysi) and connecting this database to the large collection of mutant phenotypes in the ZFIN database, the central database of the zebrafish model organism community. The evolutionary and mutant phenotypes are being described using common ontologies. The database with its web-interface, called EQSYTE (Entity-Quality System for Trait Evolution), together with the extended ontologies and data curation tools, will allow researchers to ask novel questions about the genetic and developmental regulation of evolutionary morphological transitions. Tool and database development are being guided by use cases, or driving research questions, defined by the devo-evo community. These tools are being developed under an open-source, open-development model, and in such a way that they can be used for additional biological systems in the future.
This project is a unique collaboration between evolutionary and model organism biologists including two national centers (NESCent and NCBO), the ZFIN model organism database, the Cypriniformes Tree of Life project, the DeepFin Research Coordination Network, and the morphological image databases used by the evolutionary biology community (Morphbank, MorphoBank, DigiMorph, Digital Fish Library).
The Role of Ontologies
Background
Ontologies are constrained, structured vocabularies with well defined relationships among terms. Ontologies represent the knowledge-base of a particular discipline, and provide not only a mechanism for consistent annotation of data, but also greater interoperability among people and machines. The most widely used biological ontology is the Gene Ontology, which is utilized to annotate molecular function, biological processes and subcellular localization to gene products from different organisms.
Phenotype ontologies
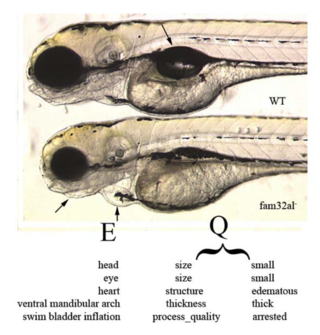
For model organisms : Approximately 500 mutant zebrafish lines (alleles) with over 660 annotated phenotypic characters from the jaw or gill arches (n=250), fins (n=210), axial skeleton (n=190) and other features (n=10) of the skeleton have been described. Researchers in the Zebrafish Information Network (ZFIN) are annotating mutant phenotypes using the zebrafish anatomy ontology and the Phenotype And Trait Ontology (PATO). PATO is a “universal” ontology of terms describing qualities (e.g. shape, color, size) that may be applied to any organism.
Anatomical ontologies
A multi-species ontology for ostariophysan fishes, The Anatomy of Ostariophysi (TAO), is being developed by expanding on the terms in the zebrafish anatomical ontology. To begin with, development of the TAO will concentrate on the skeletal system because it varies significantly across the Ostariophysi, is well-preserved in fossil specimens, and it is often the focus of morphologically-based evolutionary studies in ichthyology. The zebrafish anatomical ontology currently contains 236 skeletal system entities.
The multi-species anatomy ontology for ostariophysan fishes will be used in combination with the PATO ontology (see EQ format) to describe the naturally occurring phenotypes in non-model species (i.e. various ostariophysan fish species). We are also developing a separate catalog of homology statements for entities within TAO, so that individual investigators may select particular relationships based on evidence.
Taxonomic ontology
Together with taxonomic experts, we are developing a taxonomic ontology (based on the Catalog of Fishes) in order to relate species with particular characters and states. The taxonomic ontology will include nodes ancestral to the Ostariophysi as far back as the Vertebrata in order to associate certain anatomical terms with more inclusive clades than the Ostariophysi.
Fish Morphology
Although the comparative anatomy of fishes has been documented in the literature for several hundred years, it is not available in a computable format. A Data Curator will input morphological character data that is gleaned from the literature, culled by experts and the ichthyological community. Our goal is to input approximately 4,000 morphological features in an “EQ” format (Mabee et al. 2007a) using a combination of ontologies.
Contact
Paula Mabee (University of South Dakota) is the Principal Investigator. Co-principal investigators are Todd Vision (University of North Carolina, Chapel Hill), Monte Westerfield (University of Oregon, ZFIN), and Hilmar Lapp (NESCent) (see their contact addresses).
Acknowledgements
| This project is funded by NSF grant BDI - 0641025, and supported by the National Evolutionary Synthesis Center (NESCent), NSF #EF-0423641. This project arose from a NESCent Working Group led by Paula Mabee and Monte Westerfield, "Towards an Integrated Database for Fish Evolution." Goals and summaries of the group are archived on this wiki. |

|
News
- We will be hosting a "Needs Analysis Workshop" for this project on Sep 17-18, 2007.
- We are recruiting a Research Programmer to be in charge of data modeling, database design, middleware, and data service layer. The position will be located at NESCent and work directly with the research programmer for the user interfaces. More details are on the NESCent employment page.