Knowledgebase mockups (Nov 2009)
From phenoscape
Publications
Revision 1
Revision 1 posted on 12/16/09 by Wasila based on amphibanat feedback, Phenoscape conference call (11/20/09), and discussion with Paula (12/14/09). The Powerpoint file containing the revisions is on the webdav.
Publication results on anatomy term search pages, revision 1
List of publications, revision 1
Publication detail page, revision 1
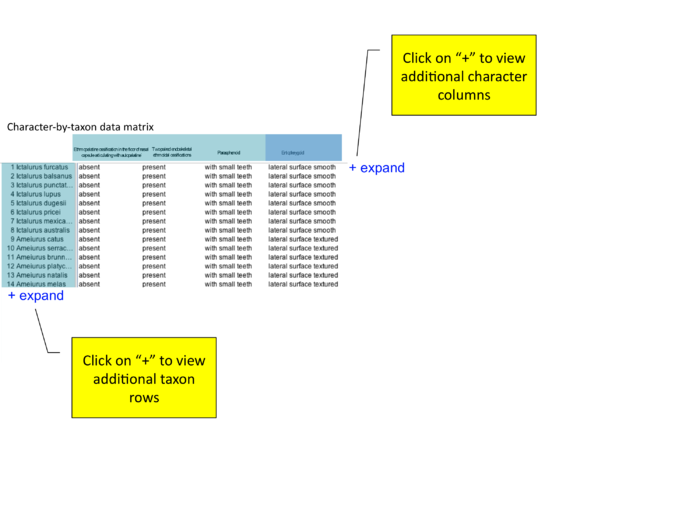
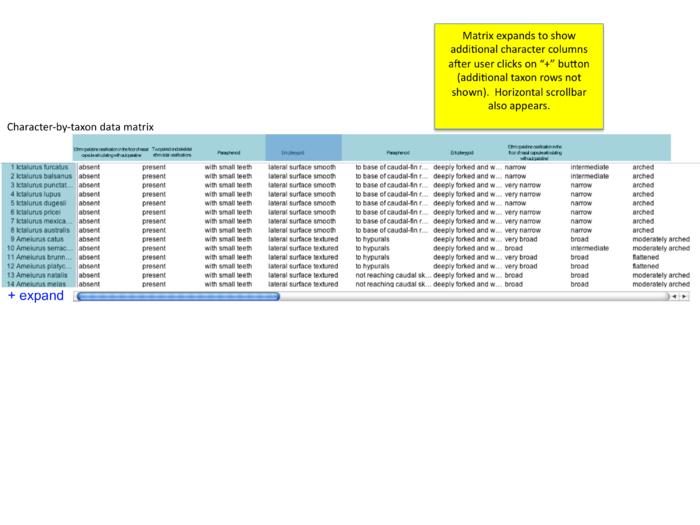
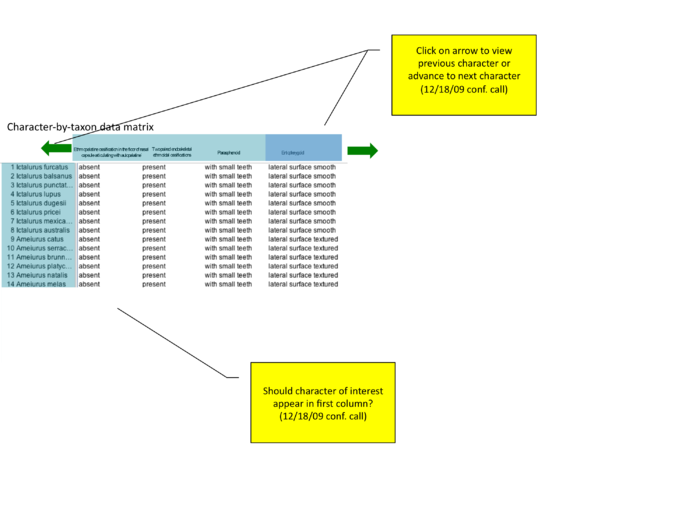
Publication detail page: viewing matrix and character of interest
from conference call 12/18/09 "how to show character of interest? should entopterygoid be first column (even if author lists as #4 or #79)? or should there be a slider or 'next' arrow that allows users to view additional characters?"
Mockups added on 12/21/09 by Wasila.
collapse/expand all button
2.
advance to next character arrow
Original mockups
Integration of publication data across site.
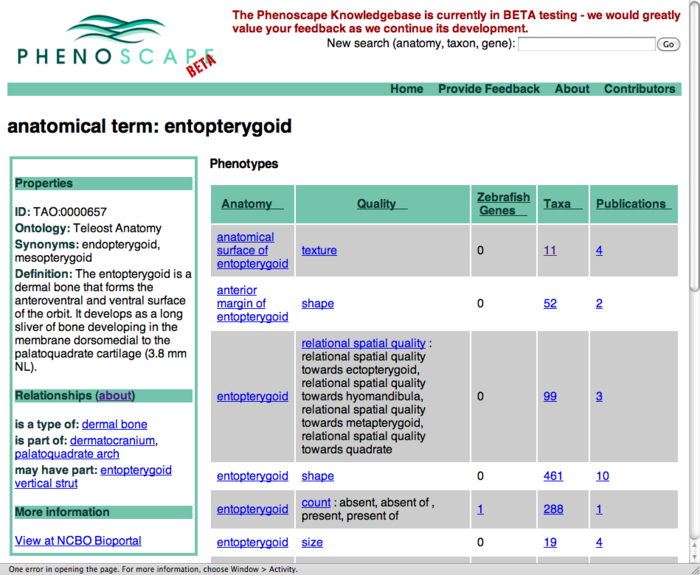
Publication results on term search pages
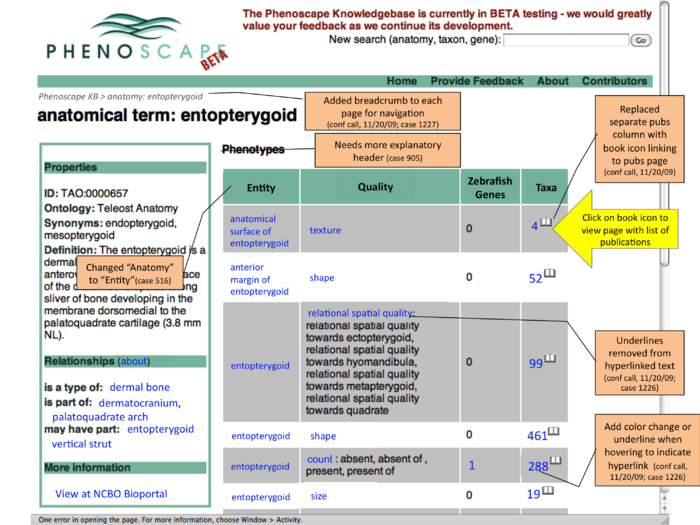
- this shows an entity page (entopterygoid) - could have similar layout on taxon and gene searches
- for each row (phenotype class) - see how many publication are linked to any EQs of that type
- click one of the count links to go to a page listing those publications
List of publications
- click one of the links to go to that publication's detail page
Publication detail page
- matrix will usually be much bigger - will be scrollable
- click column header or cell links to go to a phenotype details page showing all related EQs grouped by taxon (as on existing phenotype pages)
Comments
Tree mapping
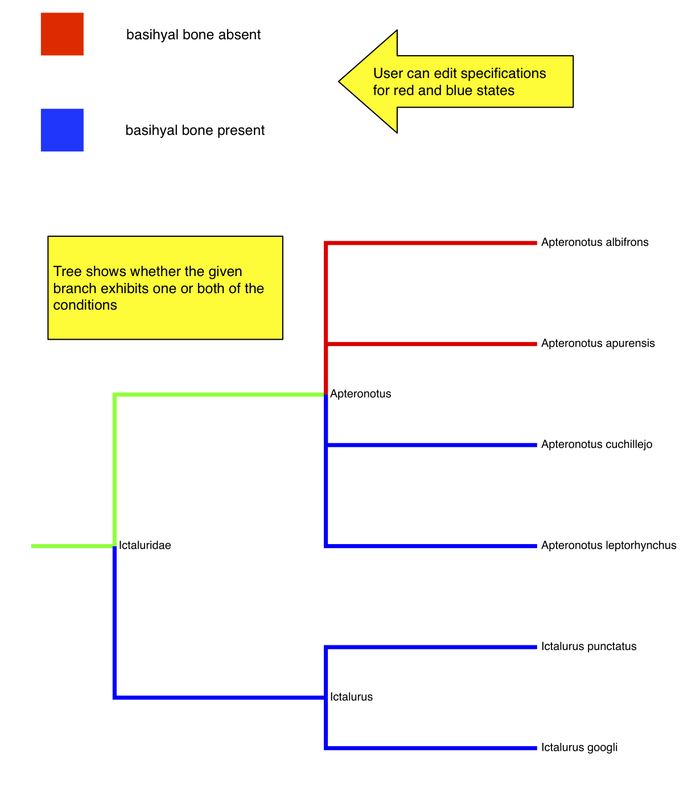
Map 2 phenotypes on tree
- user can enter phenotype specification for each "state"
- see on tree which branches exhibit one or both of the phenotypes
- Do users want the tree "pruned"? Or show branches without annotations also? could be a lot of taxa
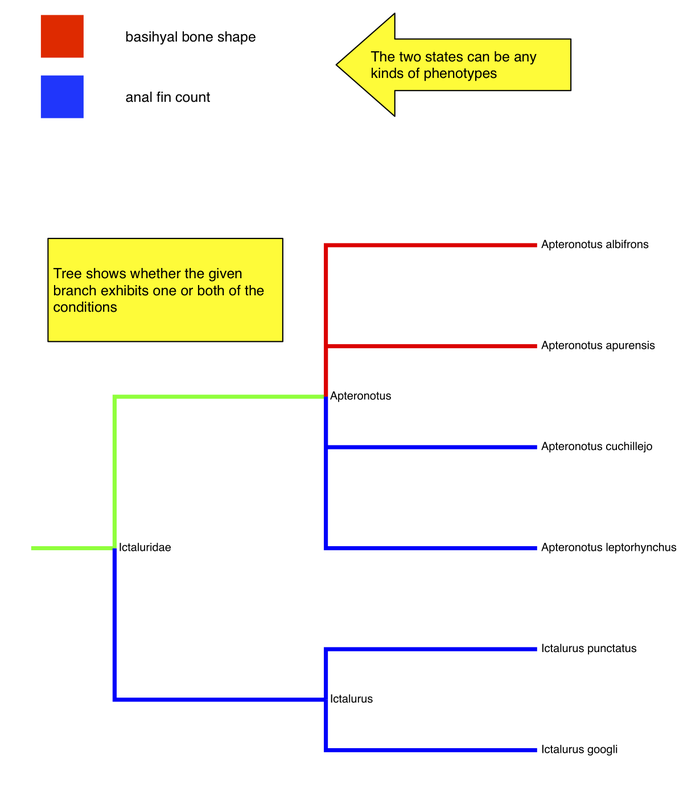
Same interface
- shows that the 2 phenotypes don't have to be obvious alternatives
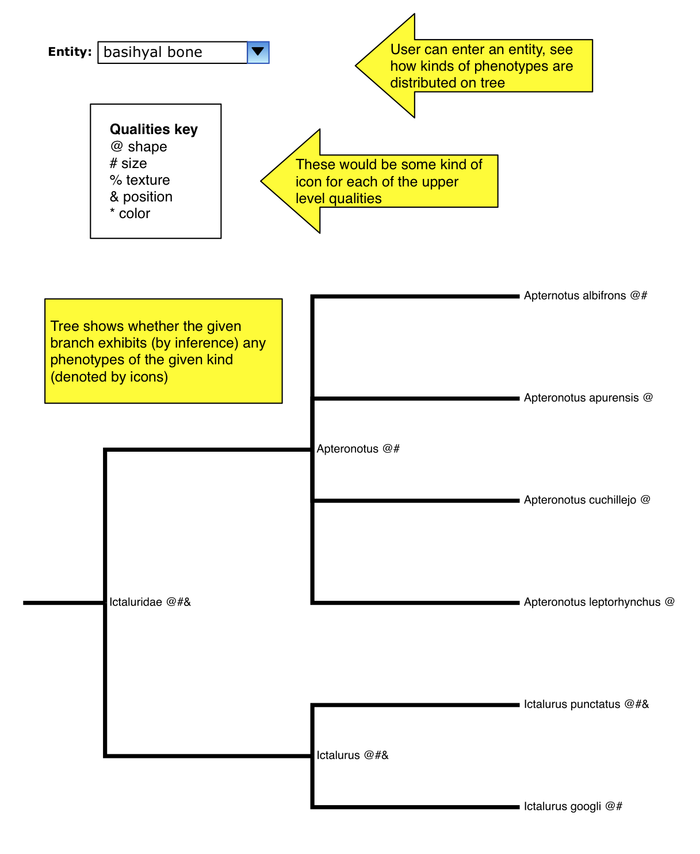
Map phenotypes of a given entity on tree
- user enters any entity term
- each of the standard upper-level PATO qualities has some icon
- each node of tree is annotated with icons for which kinds of phenotypes are exhibited by that node
- so if you just put in "bone", you could get a "shape" icon at a node for any bone shape phenotype (basihyal bone round, vertebra 1 flat, etc.)
Comments
Phenotype queries
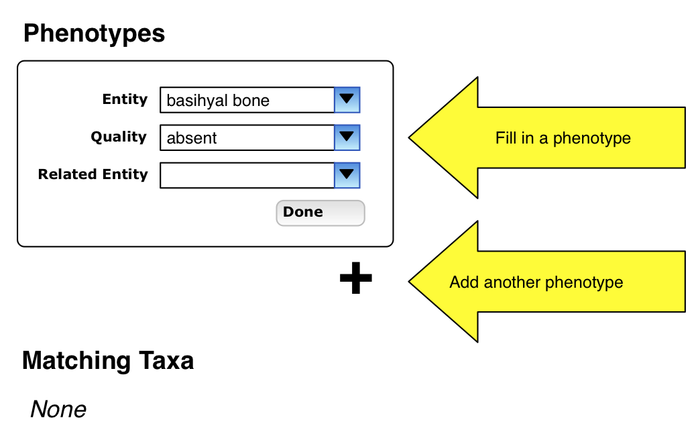
Entry of phenotype specification
- user enters a phenotype specification, presses Done
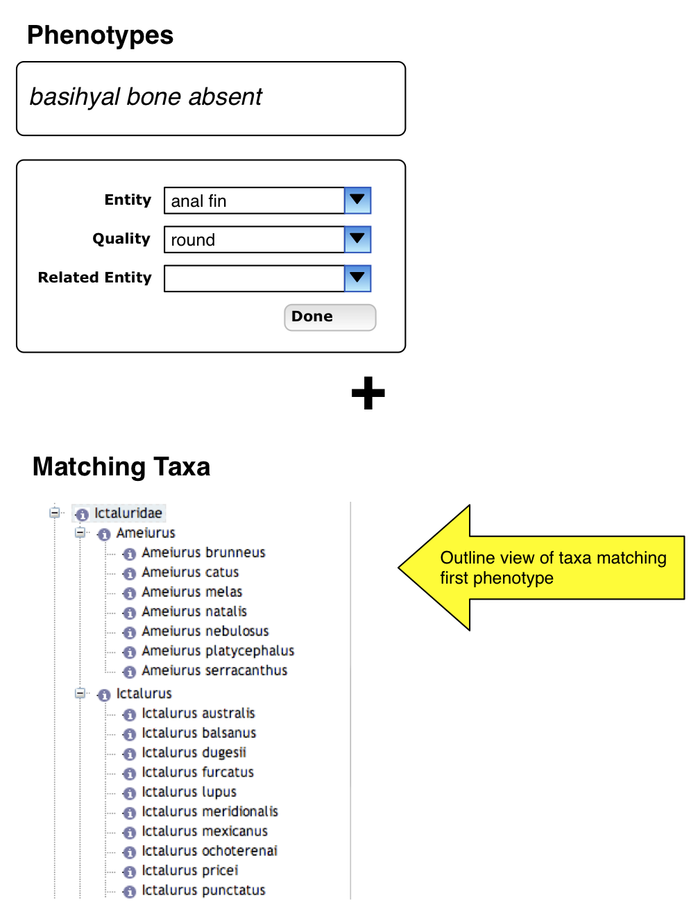
Additional phenotype specification
- bottom of page shows taxa which exhibit first phenotype
- user pressed "+" under first phenotype and can enter another
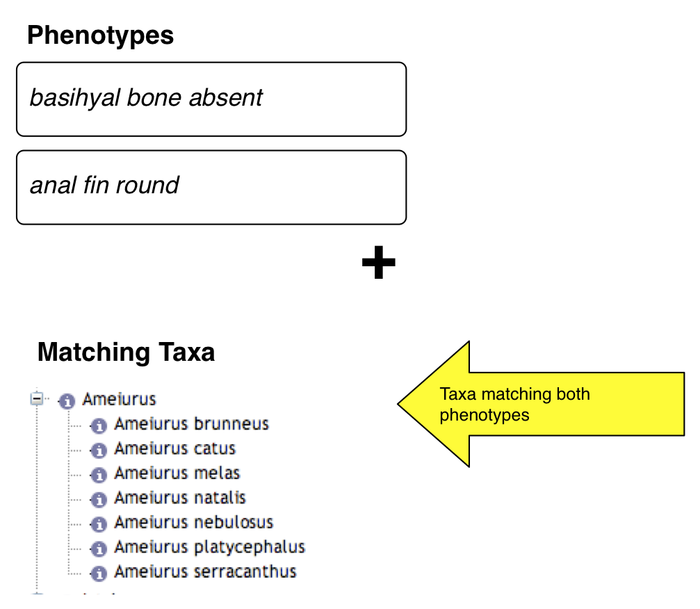
Results
- taxa exhibiting both phenotypes are shown
Comments
What should happen upon clicking taxon names? --jim 11:38, 18 November 2009 (EST)
- go to taxon's page?
- go to phenotype listing for that taxon and given phenotypes?