This tutorial is designed to introduce users to exploring phenotypic data in the Phenoscape Knowledgebase (KB). The current release includes data from over 50 comparative publications, describing mainly ostariophysan fishes (for example, catfish, minnows, loaches, characins, and knifefishes), as well as developmental genetic data imported from ZFIN.
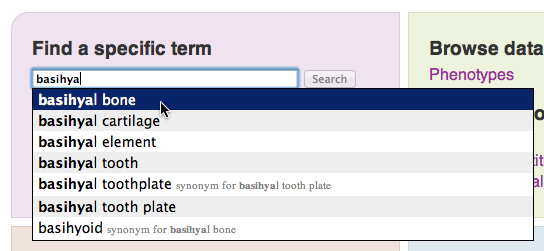
The homepage provides several different entry points into the KB. We’ll start by using the term search field to find data related to a structure of interest, such as the “basihyal bone”. The KB consists of structured data in the form of related ontology terms. So the search field is used to find a specific term from the vocabulary known to the KB. An autocomplete menu will pop up once you have typed 3 characters. Enter “basihyal bone” into the search field and click on it once it appears in the popup menu.
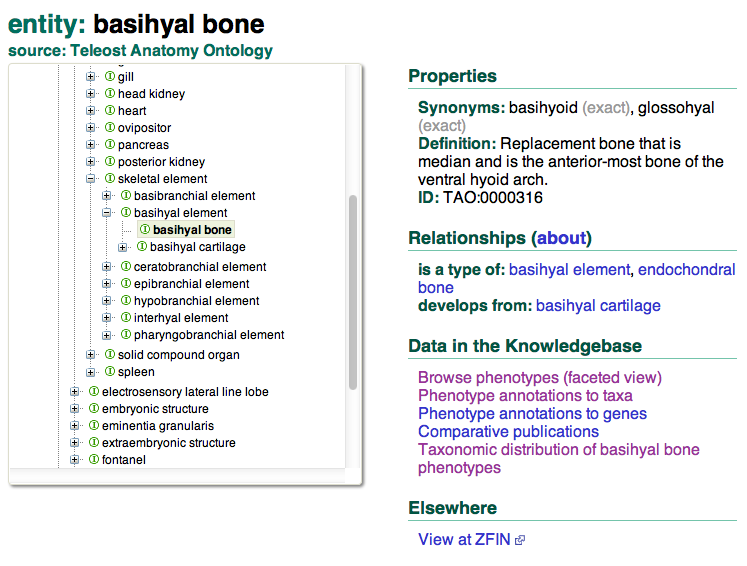
Pressing the Search button will take you to the term page entry for this structure: you can view the placement of this term within the anatomy ontology hierarchy, metadata such as synonym labels and a textual definition, semantic information such as logical relationships to other concepts, and links to data pages in the KB focused on the basihyal bone. To get an overview of the data in the KB about the basihyal bone, we’ll follow the link Browse phenotypes (faceted view).

This takes us to a faceted browsing page—faceted browsing lists
phenotypes that have been used in annotations within the KB, filtered by
the facets on the left side of the page: entity, quality, taxon, or
gene. The entity facet is already restricted to basihyal bone. By
looking at the quality facet, we can see the categories of qualities
associated with basihyal bone in the KB, and the number of different
phenotypes for each. Click on
morphology
in the quality facet to further limit the data to phenotypes about
basihyal bone morphology. The results table lists phenotypes that have
been used in annotations of taxonomic characters or genes.  To explore basihyal bone morphology in more detail, click the link above
the facets panel which says View these results as a phenotypes
query.
To explore basihyal bone morphology in more detail, click the link above
the facets panel which says View these results as a phenotypes
query.
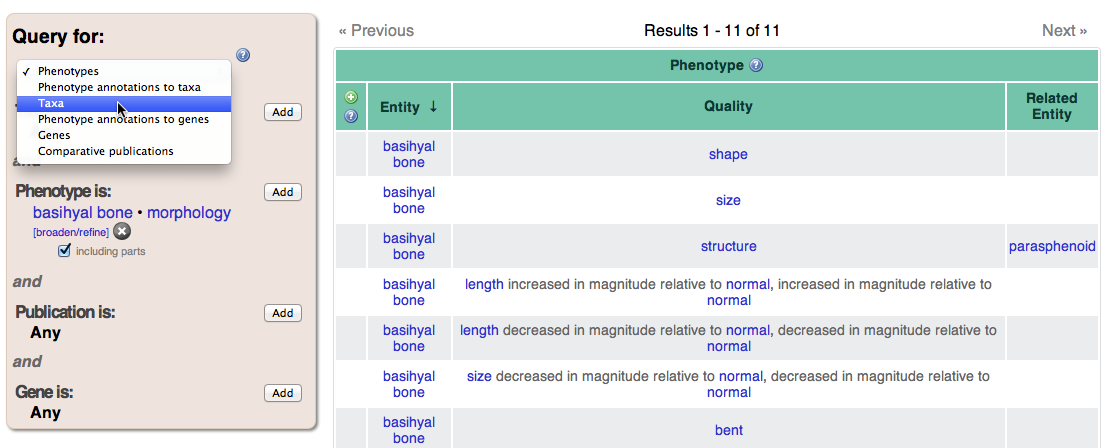
Query pages have a query panel on the left and results table to the right. After following the previous link, the query panel will be pre-filled with a phenotype consisting of the entity “basihyal bone” and the quality “morphology”. The type of result we are querying for can be changed using the dropdown menu at the top of the query panel. It should currently display Phenotypes; change it to Taxa to view taxa with data for basihyal bone morphology in the KB. In order to further refine the search, click Add next to the Taxon section of the query panel. In the resulting dialog, you can enter any taxon (try “Gymnotiformes”) to see only taxa within that group. Again using the drop down menu at the top of the query panel, you can change the query type to Phenotype annotations to taxa to view specific phenotypic annotations. The Source column provides a link to the original character state descriptions that have been annotated with this phenotype.
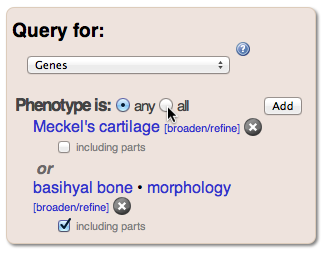
In addition to viewing comparative data, we can also view related developmental genetic annotations from zebrafish. Using the Query for: dropdown menu, change the query type to Phenotype annotations to genes. The resulting table will display ZFIN annotations for genes affecting basihyal bone morphology. Source information for the zebrafish phenotype can be obtained from ZFIN by clicking the icon in the Source column and following the link. Some of these genes have multiple annotations matching the search criteria. To get a list of unique genes affecting basihyal morphology, change the result-type dropdown to Genes. The query panel allows us to add multiple criteria to the search: click Add within the Phenotype section and enter “Meckel’s cartilage” as the entity in the Choose Phenotype dialog. Initially the query will show genes that affect either Meckel’s cartilage or basihyal, but if you change any to all in the phenotype section, you will see only genes that affect both structures. Note: depending on the query type, intersections (“all”) are not always available. Try changing the query result type, using the drop down menu, to see what options are available.
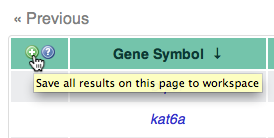
Using the Workspace, it is possible to save results obtained via
queries to be used as input to further queries. For example, we can
learn more about the genes affecting both Meckel’s cartilage and the
basihyal bone: in the upper left corner of the results table, click the
green + icon to add them all to the Workspace. Then click Workspace
near the top of the page to use your saved
items. At first the genes we
saved are grayed out, since they aren’t useful for the initially
selected query. 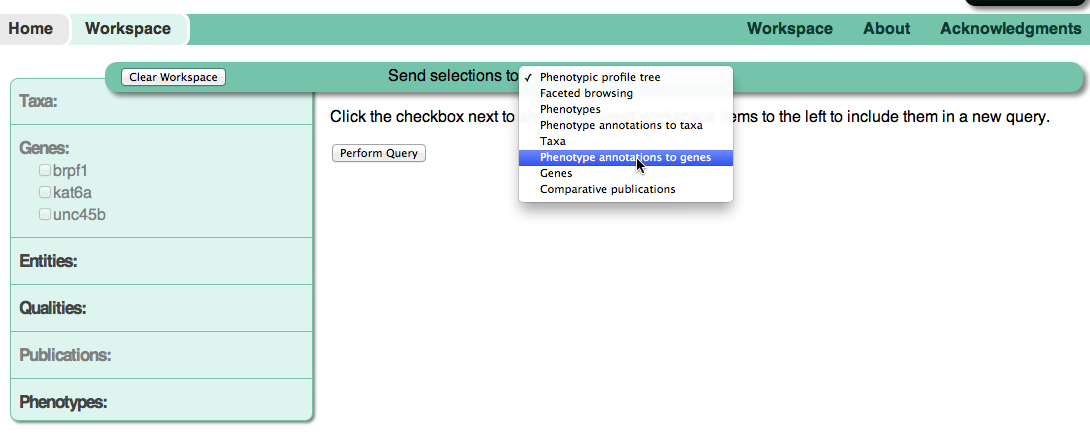 Using the Send selections to: dropdown menu, change the query type to
Phenotype annotations to genes. Then check the box next to each gene
and press the Perform Query button. You’ll now be able to view the
annotations for all of these
genes.
Using the Send selections to: dropdown menu, change the query type to
Phenotype annotations to genes. Then check the box next to each gene
and press the Perform Query button. You’ll now be able to view the
annotations for all of these
genes.
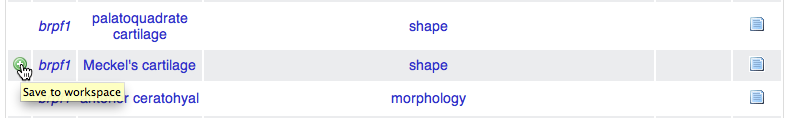
The KB contains information about the phylogenetic publications which
have been annotated by Phenoscape. We can use the Workspace to navigate
to some of interest. Within the previous results
table,
we can see that the brpf1 gene affects Meckel’s cartilage shape. Click
the + icon in the far left column of that result row to add that
phenotype to the Workspace (the icon appears when your mouse hovers over
the cell). Return to the workspace
page and change the query type to
Comparative publications using the dropdown menu. 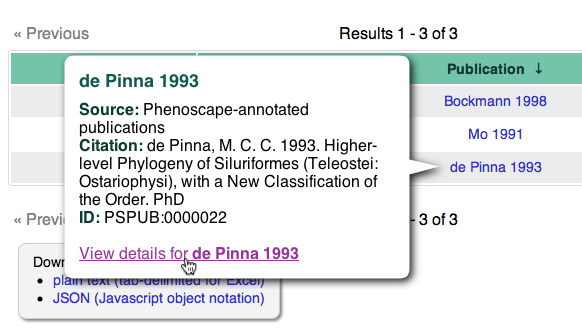 Check the box next to “Meckel’s cartilage shape” in the Phenotypes
section, then press the Perform Query button to see all the
publications in the KB containing characters relating to the shape of
Meckel’s
cartilage.
You can click on one of the publication names, such as “de Pinna 1993”,
to see a popup with its full citation. Follow the View details for de
Pinna 1993 link to see the detail page for this
publication,
including the abstract, original character matrix, and a list of
examined specimens.
Check the box next to “Meckel’s cartilage shape” in the Phenotypes
section, then press the Perform Query button to see all the
publications in the KB containing characters relating to the shape of
Meckel’s
cartilage.
You can click on one of the publication names, such as “de Pinna 1993”,
to see a popup with its full citation. Follow the View details for de
Pinna 1993 link to see the detail page for this
publication,
including the abstract, original character matrix, and a list of
examined specimens.
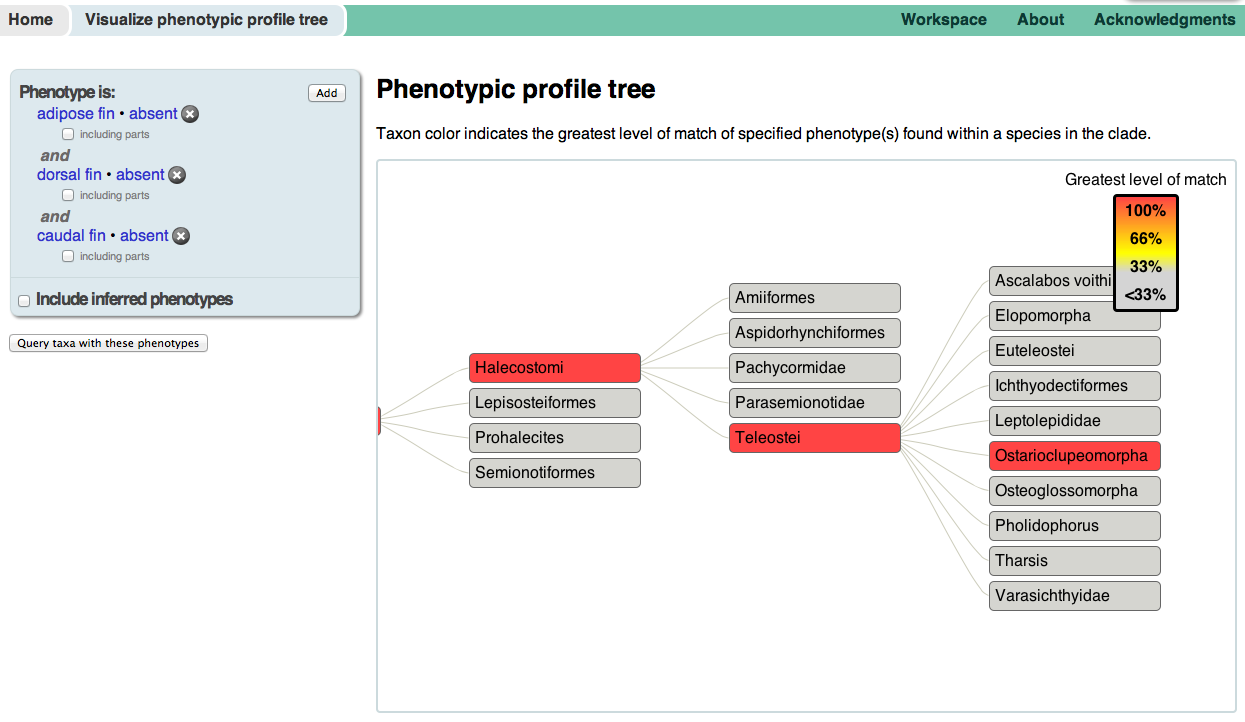
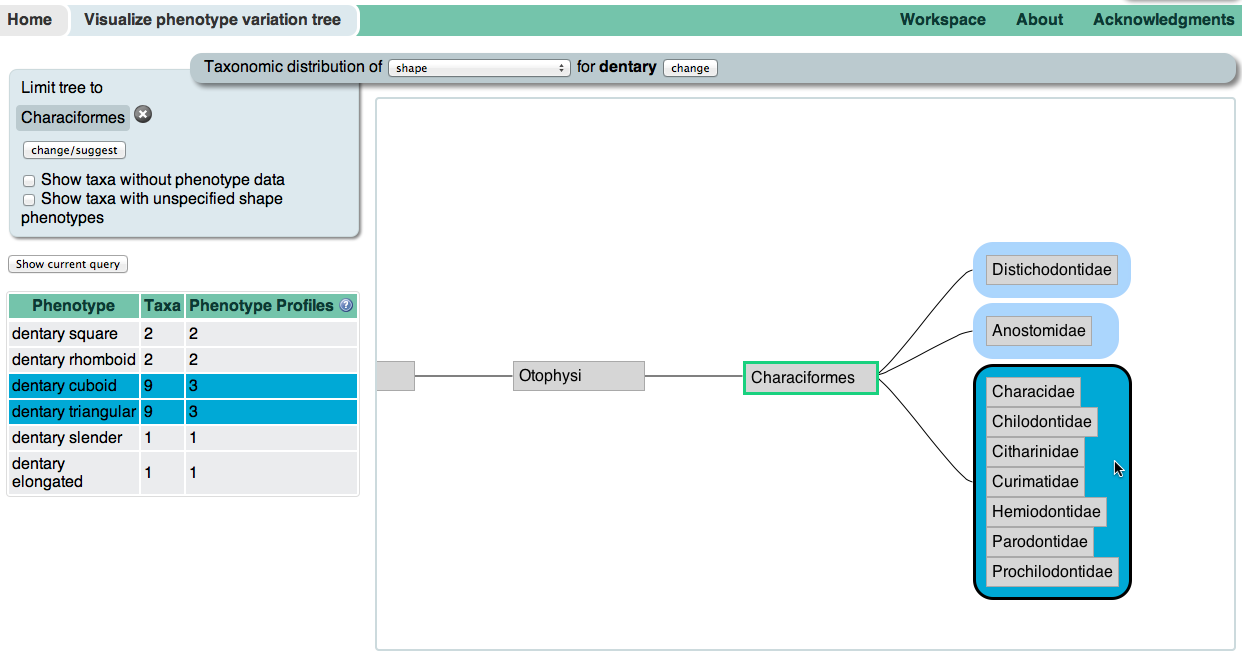
Two of the more experimental interfaces to the KB use the organismal taxonomy to display and browse phenotypic annotations. Both can be accessed from the Visualize data panel on the homepage. The Phenotypic profile tree allows you to enter multiple phenotypes, and then browse the taxonomy for branches in which some or all of the phenotypes co-occur. For example, we can find browse for taxa which are missing the adipose fin, dorsal fin, and caudal fin. The Phenotypic variation tree allows you to see the taxonomic distribution of all phenotypes matching a specification, such as dentary bone shape. Hover over a taxon in the tree to see which phenotypes in the table to the left occur there, and hover over a phenotype to see in which taxa the phenotype occurs.
 Knowledgebase data can be downloaded in bulk—look for the Download
links following data tables on each page. JSON is a structured text
format useful to programmers. The plain-text output is tab-delimited and
can be viewed within Microsoft Excel. Please contact us if you are
interested in a different format (such as RDF) or would like a dump of
all the data.
Knowledgebase data can be downloaded in bulk—look for the Download
links following data tables on each page. JSON is a structured text
format useful to programmers. The plain-text output is tab-delimited and
can be viewed within Microsoft Excel. Please contact us if you are
interested in a different format (such as RDF) or would like a dump of
all the data.
Please contact us with suggestions on how to make this interface work better for you. At the top of each page of the KB, there is a prominent Feedback button. You can also send an email to help@phenoscape.org.