Our overall objective is to create a scalable infrastructure that enables linking descriptive phenotype observations across different fields of biology by the semantic similarity of their free-text descriptions. In other words, we are trying to make descriptive observations amenable to large-scale computation so that they can be subjected to computational data integration and knowledge discovery techniques in ways similarly powerful as the techniques we are used to for numeric, quantitative observations.
Our approach to accomplish this centers on transforming descriptive observations from the natural language text form in which they are typically reported, to fully computable logic expressions that utilize terms from shared ontologies. We create these expressions (which we also call “annotations”) for evolutionary phenotypes reported in the systematics literature, typically in the form of character state matrices. We use the Entity-Quality (EQ) formalism to compose these expressions, which was initially conceived for making biomedical and mutant model organism phenotype observations interoperable.
We combine the EQ annotations we create for evolutionary phenotypes with the EQ annotations created for the myriad of phenotypes observed for mutant model organisms in an integrated knowledgebase (essentially a triple-store). We then apply Description Logic-reasoning to evaluate which evolutionary phenotype transitions can be inferred as semantically similar to which mutant model organism phenotypes, and vice versa. Since the genetic cause of a mutant phenotype is usually known, the links between evolutionary and mutant phenotypes identified in this way can be used to construct testable hypotheses about the genetic correlates or causes of evolutionary transitions.
 In a previous project, titled
Linking
Evolution to Genomics Using Phenotype Ontologies, we developed a
working prototype as a successful proof-of-concept, using
teleost fishes for
evolutionary phenotypes and the zebrafish model
organism as a source of mutant phenotypes. Here, we
aim to make the components of the prototype, including tools and
workflows, sufficiently scalable so that they are adequate for the much
more extensive volume and more diverse nature of skeletal phenotypes
across all vertebrates, fossil and modern. Specifically, our aims
encompass the following:
In a previous project, titled
Linking
Evolution to Genomics Using Phenotype Ontologies, we developed a
working prototype as a successful proof-of-concept, using
teleost fishes for
evolutionary phenotypes and the zebrafish model
organism as a source of mutant phenotypes. Here, we
aim to make the components of the prototype, including tools and
workflows, sufficiently scalable so that they are adequate for the much
more extensive volume and more diverse nature of skeletal phenotypes
across all vertebrates, fossil and modern. Specifically, our aims
encompass the following:
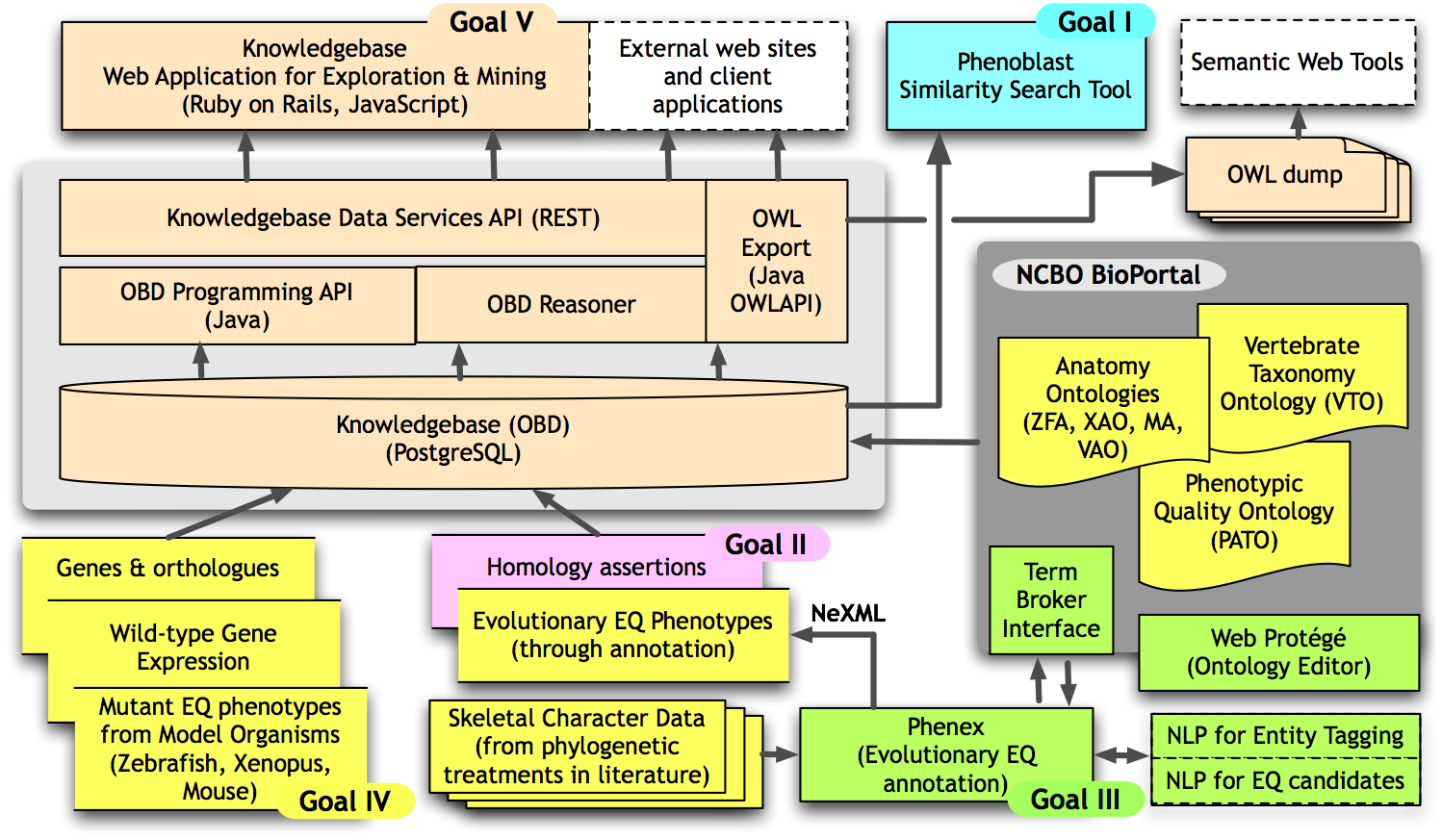
In addition to a web-based interface, we will make all data, including the integrated knowledgebase, available in the Web Ontology Language (OWL), so that other researchers can reuse the data in as many ways as possible.
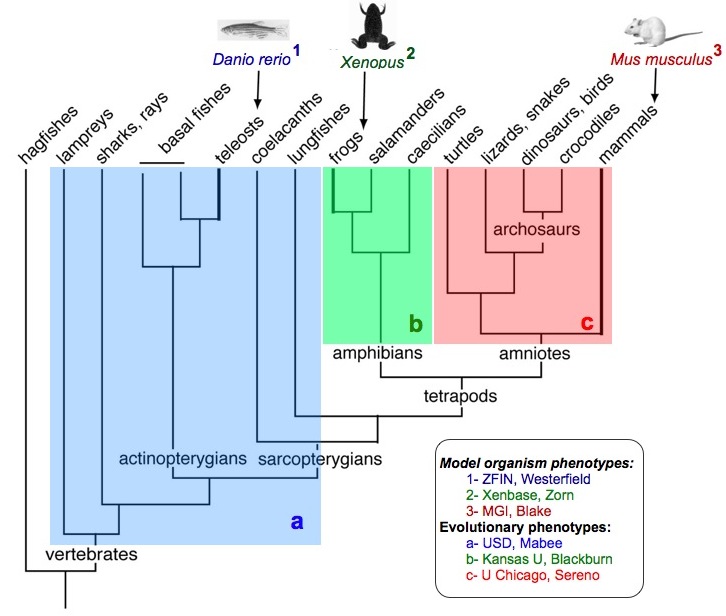
The evolution of limbs from fins is arguably one of the most well studied transitions in vertebrate history. The genes involved in positioning, growth, and patterning of the fin and limb at various stages are well-known and documented in the vertebrate model organism databases ZFIN, Xenbase, and MGI; changes in skeletal morphology and corresponding assertions of homology are well-documented in the comparative morphological literature. Bringing together the genetic, developmental, morphological and evolutionary data in the Phenoscape Knowledgebase will provide an ideal test bed for judging the reliability of candidate gene predictions and the application of homology logic.
Outreach activities include:
Paula Mabee (University of South Dakota) and Todd Vision (University of North Carolina Chapel Hill, National Evolutionary Synthesis Center) are the Principal Investigators of this project. Co-principal investigators are David Blackburn (California Academy of Sciences), Judith Blake (Mouse Genome Informatics, Jackson Laboratories), Hilmar Lapp (National Evolutionary Synthesis Center), Paul Sereno (University of Chicago), Monte Westerfield (ZFIN, University of Oregon), and Aaron Zorn (Xenbase, Cincinnati Children’s Hospital Medical Center) (see their contact addresses).