The Phenoscape data repository is a relational database, which holds phenotypic data from the model organism Danio Rerio (Zebrafish) and the evolutionary organisms belong to the clade of Ostariophysi. This page describes the schema of this data repository and outlines the methods to load and query this data repository.
The Phenoscape data repository has been implemented as a PostgreSQL relational database, and at present is housed on a dedicated database server.
The schema of the Phenoscape data repository is based upon the Open Biomedical Database (OBD) data format developed at the Berkeley Bioinformatics Open-source Projects (BBOP). OBD is based upon the Resource Description Framework (RDF) format for capturing metadata about Web (and Semantic Web) resources such as Web pages and Web services.
The philosophy of OBD is to represent every conceptual entity, be it a type or a token (synonymously a class or an object, or a concept or an instance) or a relation definition, as a Node. Instances of relations between these nodes are represented as Statements, specifically Link Statements. OBD also allows for reification, which is vital to the life sciences with their emphasis on evidence codes and attributions (provenance). For this purpose, OBD provides Literal Statements (and Annotation Statements) to capture metadata about Nodes and Link Statements, such as the source publication, evidence codes, specimens used, and so forth.
Two relational tables are central to the schema of the Phenoscape data repository. These are: LINK and NODE. The SQL commands for the creation of these tables (and the others) can be found at this Phenoscape Sourceforge page.
The NODE table contains information about every concept such as its unique identifier, label, and source ontology. The NODE table contains this information about concepts extracted from the source Ontologies. In addition, it also holds information about scientific publications (in a rudimentary format which will be improved upon soon), the ontologies themselves, and the representation of phenotypes from the ZFIN and NeXML databases. It will be augmented in the future to hold information about collection specimens. The NODE table adds a unique identifier (generated from a sequence) of its own to every row. An excerpt of the row from the NODE table for the Gymnotiformes term is shown below
The LINK table contains rows which represent Statements which link the Nodes to one another, and also the metadata about these Nodes. The excerpt below shows some of the rows in the LINK table about the Gymnotiformes term
In simple terms, a sub species of Gymnotiformes is displayed by this Statement as shown in the triple below
Similarly, The third row in the display shows that Gymnotiformes is an Otophysian as shown below
The Phenoscape data repository also generates several views from the tables. These views are used in querying the database, some of which are part of the OBDAPI
Stored procedures are used in populating the database with defined terms from the ontologies, and with phenotypic descriptions obtained from curators. In addition, they are also used in generating inferences from the asserted data. In the future, stored procedures may be used as necessary for speedier data retrieval.
The repository will be periodically refreshed to include the latest ontology definitions and curated data. At present, curated data is obtained from two different source which are:
A complete database refresh using the Phenoscape data loader can be started off by running the “refresh-database” target in the Ant build file in the ‘Phenoscape’ folder of the OBDAPI project.
Queries have been implemented for retrieving phenotype information (summaries and details), homology information, summaries of search terms, metadata about phenotype assertions, and auto complete suggestions for search terms as they are being entered. Data retrieved by these queries are accessed by the various Phenoscape data services. The details about these queries are presented here.
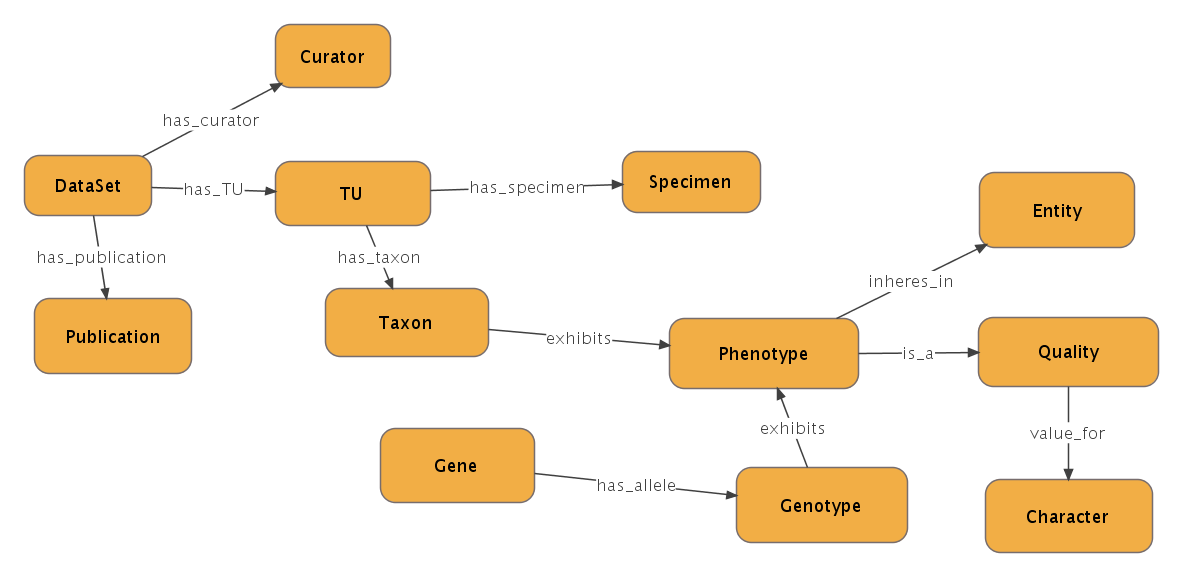
This section discusses the various entities and binary directed links between these entities, which are leveraged by the database queries. Assertions about the model organism (from ZFIN) and the evolutionary species are converted into the exhibits link specified in (1) below. Note the right hand side of the exhibits link. It is a post composition of an Entity and a Quality, which makes up a description of a phenotype.
The post composed phenotype is related to its components by the is_a and the inheres_in relation as shown in (2) and (3) below
The Quality is related to a Character by an inferred value_for relation as shown in (4)
An example should make this clearer. Consider the statement, “In Siluriformes, the shape of the dorsal surface of the basihyal bone is flat or convex” from [Albert, 2001]. This statement can be represented as in (1ex) below. Note the similar form to (1). Siluriformes is the taxon, flat is the quality, and basihyal bone is the entity.
Now the post composed phenotype is related to its entity and quality components as in (2ex) and (3ex). Note the similarity to (2) and (3)
Finally, the quality ‘flat’ is related to the character ‘shape’ by (4ex). Note that ‘flat’ is just one of the values for ‘shape’. Other values my be ‘rounded’, ‘curved’, etc.
Moving on, the database also stores provenance information (metadata) about the assertion that Siluriformes exhibits flat basihyal bones. AT the very minimum, we need to know the publication from which the assertion was extracted. If the curators have specifically cited the text from the publication which forms the basis of their assertion, we need to know that as well.
The database provides a handle to access this metadata from the assertion itself. The LINK table includes a reiflink_node_id attribute, from which publication, curator names, character and state text, and all other relevant metadata can be accessed. Without going into more database specific details, conceptually the statement (1ex) is linked to a reification identifer, which is linked to the actual metadata. Transparently, the statement (1ex) can be linked with a publication as shown in (5ex) below. The linkage to the other facets of the metadata is done similarly.
The schema of the relations is shown below
Queries used in the Phenoscape data services module were found to be intolerably slow in returning, esp. when asked to retrieve and summarize annotation data for genes and teleost species. The slow times in query execution were primarily due to the large numbers of JOINs in them, and the extensive volume of data, which needed to be processed in various facets of the query execution plan.
To address this issue, it was decided to create summaries of the annotations in the database in simple data warehouse tables. New queries which were tested on these summary tables executed much faster, having dispensed with the numerous JOINs between the NODE and LINK tables, aliased several times over.
title=” The data warehouse schema “> The data warehouse schema </a>
The data warehouse has been designed with the intent of maximizing the efficiency of queries executed on the Phenoscape knowledge base. For phenotype queries, we need to know the phenotype in question, the taxa or genes which are associated with that phenotype, as well as the entity and quality associated with that phenotype. We also need to find the character, which the quality is associated with. For example if the quality is reduced number of, the character in question would be count.
To effectively execute this query, the phenotype centric model of the data warehouse is designed as follows (concepts and attributes are capitalized). A taxon or gene may be associated with one or more PHENOTYPE(s) and a PHENOTYPE may be associated with one or more genes or taxa. A PHENOTYPE is associated with exactly one ENTITY and one QUALITY. A QUALITY may be associated with one or more PHENOTYPE(s). Further, a QUALITY is associated with exactly one CHARACTER, which is a QUALITY as well.
For queries for provenance data about taxon to phenotype assertions, we need to find the publication the assertion is extracted from, the specific text from the publication about character and state, as well as the curators’ comments about the assertion.
To effectively execute these ‘metadata’ queries, the provenance data is modeled as an association attribute. For every instance of the association between a TAXON and a PHENOYPE, we capture CHARACTER. STATE, CURATORS, and PUBLICATION. The PUBLICATION entity with all its attributes is linked to the REIF entity, which is the link to the metadata of the TAXON and PHENOTYPE.
This data warehouse can be reduced to the logical schema shown below
| Gene | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene_id {PK} | Gene_Uid | Gene_Label | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene_Alias | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene_id{FK} | Alias | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Genotype | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Genotype_id{PK} | Genotype_Uid | Genotype_Label | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxon | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxon_id {PK} | Taxon_Uid | Taxon_Label | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxon_Alias | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxon_id {FK} | Alias | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxon_Is_A_Taxon | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxon_id{FK} | Taxon_id{FK} | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Entity | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Entity_Id {PK} | Entity_Uid | Entity_Label | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Entity_Is_A_Entity | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Entity_Id {FK} | Entity_Id {FK} | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Entity_Part_Of_Entity | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Entity_Id {FK} | Entity_Id {FK} | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Quality | |||||||||||||||||||||||||||||||||||||||||||||||||||||
| Quality_Id {PK} | Quality_Uid | Quality_Label | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Phenotype | |||||||||||||||||||||||||||||||||||||||
| Phenotype_Id {PK} | Phenotype_Uid | Inheres_In_Entity_id {FK} | Towards_Entity_id {FK} | Is_A_Quality_id {FK} | Is_A_Character_id {FK} | Has_count | |||||||||||||||||||||||||||||||||
| Gene_Genotype_Phenotype | |||||||||||||||||||||||||||||||||
| Gene_Id {FK} | Genotype_Id {FK} | Phenotype_Id {FK} | |||||||||||||||||||||||||||||||
| Taxon_Phenotype | |||||||||||||||||||||||||||
| Taxon_Id {FK} | Phenotype_Id {FK} | Reif_Id {FK} | |||||||||||||||||||||||||
| Taxon_Phenotype_Metadata | |||||||||||||||||
| Reif_Id {PK} | Character_Text | State_Text | Curators | Curator_Comments | |||||||||||||
| Publication | ||||||
| Publication {PK} | Primary_Title | Secondary_Title | Pages | Volume | Abstract | Year |
| Publication_Reif_id | |
| Publication {FK} | Reif_Id {FK} |