Difference between revisions of "Matrix annotation workflow"
(→Character/character state editing) |
(→Overview) |
||
| (7 intermediate revisions by 2 users not shown) | |||
| Line 97: | Line 97: | ||
* Choose TTO term for each taxon. | * Choose TTO term for each taxon. | ||
* Enter list of specimens for each taxon, choosing museum code from fish collections ontology and entering free-text ID. | * Enter list of specimens for each taxon, choosing museum code from fish collections ontology and entering free-text ID. | ||
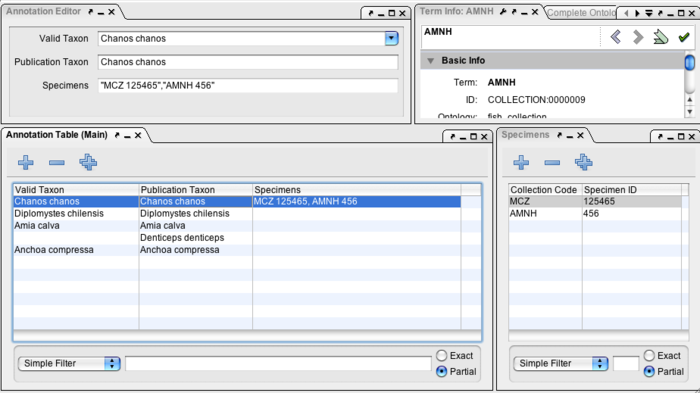
| + | [[Image:taxon_editing.png|center|700px|thumb|Taxon list editing is essentially unchanged from previous versions of Phenote. However, the list of free-text Publication Taxa can be viewed and edited in Mesquite as a Taxa block. The TTO identifier and list of specimens will be stored but invisible in Mesquite.]] | ||
====Character/character state editing==== | ====Character/character state editing==== | ||
| Line 104: | Line 105: | ||
[[Image:vomer_shape.png|center|700px|thumb|The top of the panel shows the list of characters, as can also be viewed in Mesquite. Select a character to see its possible states. Select a state to view and enter EQ phenotype statements characterizing that state. This is a mockup only - the top half would likely be in its own new moveable panel in Phenote, while the bottom half would be a standard moveable Phenote annotation table.]] | [[Image:vomer_shape.png|center|700px|thumb|The top of the panel shows the list of characters, as can also be viewed in Mesquite. Select a character to see its possible states. Select a state to view and enter EQ phenotype statements characterizing that state. This is a mockup only - the top half would likely be in its own new moveable panel in Phenote, while the bottom half would be a standard moveable Phenote annotation table.]] | ||
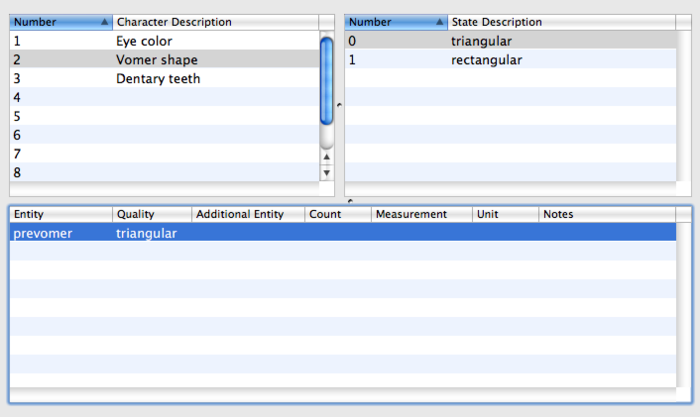
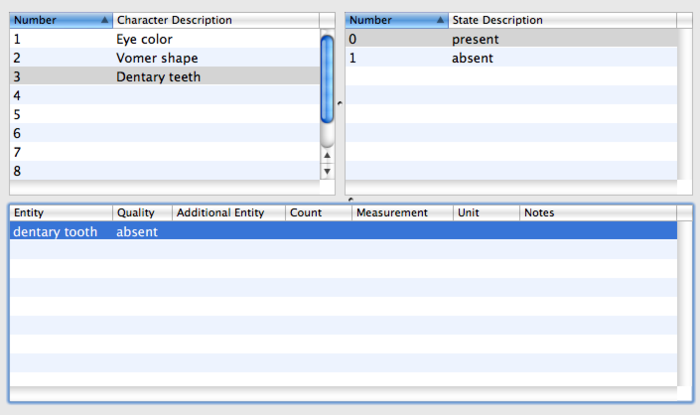
| − | [[Image:dentary_teeth.png|center|700px|thumb|Select the next character and see its states and EQ annotations. The characters and character states can also be viewed and edited in Mesquite, but the EQ annotations cannot - when the file is opened with Mesquite, the EQ annotations will be stored but invisible.]] | + | [[Image:dentary_teeth.png|center|700px|thumb|Select the next character and see its states and EQ annotations. The characters and character states can also be viewed and edited in Mesquite, but the EQ annotations cannot - when the file is opened with Mesquite, the EQ annotations will be stored but invisible. (Sorry, the EQ should be "present" - it's just a typo)]] |
== Example of integrated workflow via NEXml== | == Example of integrated workflow via NEXml== | ||
Latest revision as of 13:47, 9 September 2008
The first Phenoscape data jamboree revealed that our curation workflow implemented in Phenote could be quite cumbersome, particularly in managing the huge numbers of rows containing annotations for every taxon. Discussion after the jamboree settled on a new workflow which allows more division of effort, allowing experienced curators to focus on the description of phenotypes using EQ, while input of specimen lists and character matrices can be done separately, possibly by assistants.
Contents
Workflow activities
This workflow is divided into 3 main activities, each of which can be performed independently and in parallel.
EQ character state coding
This activity involves the translation of published character states into ontology-based EQ descriptions. This requires deep domain knowledge providing full comprehension of the published descriptions and thorough understanding of anatomy and quality ontologies. This activity can be performed independent of annotations to particular taxa - the curator focuses only on describing the character states. As such, the curator will need to deal with far fewer rows of data when working in Phenote, as compared to simultaneous taxon annotation.
Taxon and specimen lists
This activity includes entering the list of species as named in the publication ("Publication Taxon") and choosing the matching taxon term from the TTO for each. Also at this step the list of specimens studied is entered for each taxon. This activity can be easily performed by an assistant with minimal training.
Character matrix
A character matrix provides the annotation of each species with a particular character state for each described character. Many studies publish the data already in matrix form. If a matrix is not available, the curator will need to prepare one representing the data for that publication. The states in the matrix are linked to those described in "EQ character state coding" by using corresponding character and character state numbers. The taxon names in the character matrix must match Publication Taxon entries in the taxon list. This activity may also include choosing an evidence code, from the evidence code ontology, for each state assignment (each cell). If a matrix is available for a publication, this activity requires little work on the part of an experience curator.
Output from each of these activities can be merged using a non-interactive script, producing resultant taxon-specific EQ annotations.
Implementation proposal
Overview
"EQ character state coding" can continue to be performed within Phenote, being Phenote's core capability. A revised Phenoscape configuration will be created, eliminating all taxon-related columns from the annotation table, but adding a column for "character state". Curators will enter 1 or more annotation rows for each character/character state combination, with no reference to any particular taxon. The curator can enter multiple EQ's to represent any particular character state. The search filter field at the bottom of the table will be enhanced such that the curator can filter the list down to one character at a time, viewing only the EQ annotations for the states of that character. The curator will save these EQ annotations to a file for later processing.
For the time being, "Taxon and specimen lists" can continue to be entered using the current Phenote configuration. As described below, entering these will likely be incorporated into the "Character matrix" activity within Mesquite. Phenote-based taxon files will be later processed into a to-be-developed NEXUS block.
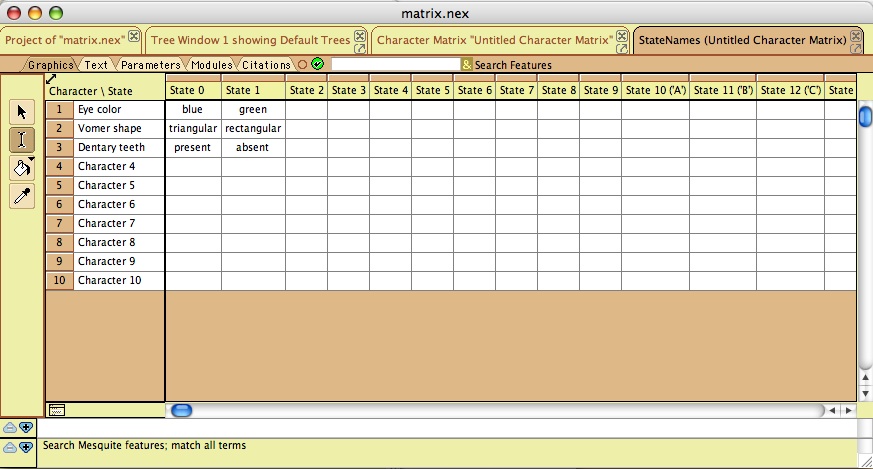
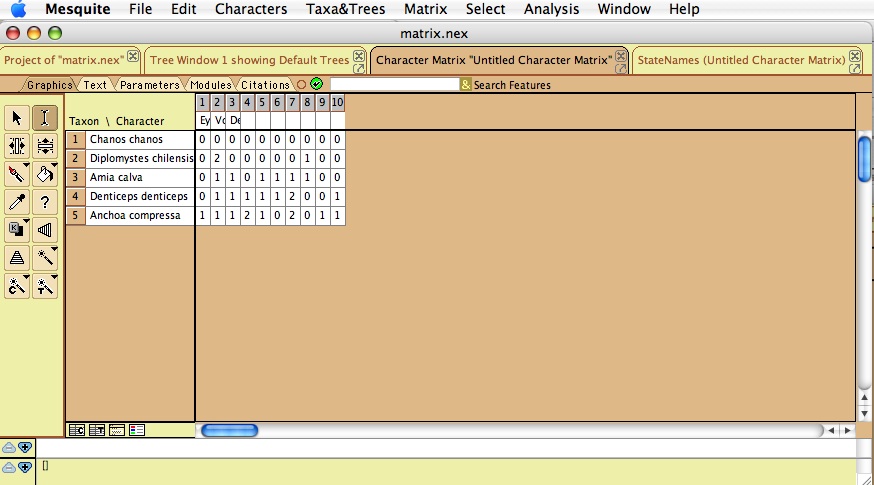
Mesquite provides a familiar and standard interface for editing evolutionary character matrices. Thus, we will use it instead of Phenote for entry of character matrices. Character columns should be ordered the same way they are numbered in the "EQ character state coding" step. Taxon names should be unabbreviated genus and species as used in the publication. Since the "Publication Taxon" is entered as part of the matrix, it would be efficient to use the Mesquite taxa list directly instead of entering a separate list as in "Taxon and specimen lists". In order to accomplish this, we will need to add some ontology capability to Mesquite, in the form of a plugin. The user could open a panel showing the taxa in the TAXA block, and choose a "Valid Taxon" from the TTO for each one. Adding such ontology capabilities will also facilitate creating an interface within Mesquite for choosing the evidence code for each matrix cell, picking from the evidence code ontology. We can also create a panel for entering specimen lists within Mesquite, eliminating a separate Taxon List step entirely.
After the matrix and taxon list information are entered in Mesquite, all ontology information (Valid Taxon, specimens, evidence codes) would be saved in one or more custom blocks in the matrix NEXUS file. This NEXUS file would be combined with the Phenote-generated EQ list to create per-taxon EQ annotations. This could happen in an external script or perhaps by loading the EQ file into Mesquite.
Requirements for Mesquite plugin
- Need to be aware of editing of TAXA block in order to keep Valid Taxon list up-to-date.
- Save and read ontology data in custom block(s).
- Make use of cell selection of matrix editor within separate evidence code panel.
Detailed Workflow 1
Detailed steps for obtaining a Publication Taxon List in NEXUS file format (creating a NEXUS file from results of Findit)
- If starting from scratch, copy taxa from selectable PDF (or re-type, otherwise: Jim why would you do this vs. just type directly into a Nexus file?) into FindIt>NEXUS web page (to be developed)
- If starting with NEXUS file, paste into FindIt>NEXUS web page (to be developed)
- FindIt>NEXUS web page will return a NEXUS file with corrected taxon names (Jim: what are "corrected taxon names"?) Some of the results from Findit will be garbage, e.g. river names, etc. that the curator will need to edit out.
Detailed steps for Character Matrix entry
- Open Mesquite or MacClade
- Open NEXUS file (this file will have Publication Taxon name list as per above)
- Enter matrix data (0,1,2, etc. for each character); save in NEXUS file
Detailed steps for Specimen entry
- Import NEXUS file into Phenote specimen list editor (NEXUS file will come from above or directly from author; if from author directly, some publication names may need to be edited (e.g. if genus is abbreviated))
- Phenote will attempt to choose a TTO taxon for each publication taxon
- Correct Phenote TTO choices as necessary
- Enter specimens for each taxon
Detailed steps for EQ entry which can happen in parallel to Taxon entry above
- Curator enters EQ phenotypes for each character state in Phenote; add free text character descriptions at this point; add Evidence codes (this will remain a Phenote function; if there is a matrix in the paper, we will assign IVS to all species, characters, and character states).
- Merge NEXUS file containing matrix with Phenote EQ file, using standalone tool (this is where we have the creation of the Phenote Character Matrix; this will be editable and we can add more taxa or characters as required)
Detailed Workflow 2
Creation of Taxon List
- Taxon List is created using the current Phenote interface and file format
- Specimen information (vouchers) are entered at this step
Creation of NEXUS file and Character Matrix entry
- Phenote exports NEXUS file with Publication Taxon names from the taxon list
- Open file in Mesquite, MacClade, or excel (possible?)
- Enter matrix data (0,1,2, etc. for each character); save in NEXUS file
Detailed steps for character matrix + specimen list merge
- Import NEXUS file into Phenote specimen list editor (NEXUS file will come from above or directly from author; if from author directly, some publication names may need to be edited (e.g. if genus is abbreviated))
- Phenote will attempt to choose a TTO taxon for each publication taxon
- Correct Phenote TTO choices as necessary
- Previously entered specimen information should appear at this step
Detailed steps for EQ entry which can happen in parallel to Taxon entry above
- Curator enters EQ phenotypes for each character state in Phenote; add free text character descriptions at this point; add Evidence codes (this will remain a Phenote function; if there is a matrix in the paper, we will assign IVS to all species, characters, and character states in bulk).
- Merge NEXUS file containing matrix with Phenote EQ file, using standalone tool (this is where we have the creation of the Phenote Character Matrix; this will be editable and we can add more taxa or characters as required)
Integrated workflow via NEXml
Ideally we could have a single file containing the character matrix and phenotype annotations, which the user could edit as needed in both Phenote and Mesquite. This should be feasible using the new standard NEXml. With this approach tasks could be performed in almost any order. Some tasks could optionally be accomplished in both Phenote and Mesquite, while some are possible in only one or the other of the programs.
File creation
The user should be able to create a new NEXml file using either Mesquite or Phenote. If an existing NEXUS matrix is available to start from, the user can open the NEXUS file in Mesquite and re-save it as NEXml.
Mesquite tasks
Taxon list editing
- Enter list of taxa, with free-text names as used in publication.
Character/character state editing
- Create and edit list of characters, with free-text for each.
- Create and edit list of states for each character, with free-text for each.
Matrix editing
- Choose appropriate character state for each taxon in matrix.
Tree editing
- Create and edit a phylogeny containing taxa from the taxon list. This tree can be saved within the NEXml file. Note, the new data-entry workflow contains no use cases involving phylogenies.
Phenote tasks
Taxon list editing
- Enter list of taxa, with free-text names as used in publication.
- Choose TTO term for each taxon.
- Enter list of specimens for each taxon, choosing museum code from fish collections ontology and entering free-text ID.
Character/character state editing
- Create and edit list of characters, with free-text for each.
- Create and edit list of states for each character, with free-text for each.
- Created and edit 1 or more EQ phenotype statements for each character state.
Example of integrated workflow via NEXml
Phenote: creation of Taxon List
- Taxon List is created using the current Phenote interface
- Valid taxon, publication taxon, and specimen information (vouchers) are entered at this step
- File is saved in NEXml format
- Note: if NEXUS file of character x taxon matrix is available, then workflow begins in Mesquite: NEXUS file needs to be converted to NEXml,and taxon names might need editing if not recorded as binomials. The NEXml file is then opened in Phenote, and Valid Taxon names are autocompleted (if that's a possible feature, otherwise chosen manually)
Mesquite: creation of Character Matrix
- Open NEXml file in Mesquite
- Display matrix view panel (only Publication Taxon names are visible because TTO is not displayed in Mesquite)
- Enter matrix data (0,1,2, etc. for each character);
- Free text character and state descriptions can be added at this point
- Save file
Phenote: EQ entry
- Open NEXml file in Phenote
- Free text character and state descriptions can also be added at this point
- Curator enters EQ phenotypes for each character state
Open issues
- Assignment of evidence codes: one idea is to enter evidence codes in a taxon x character matrix viewer in Phenote, with cells linked to the Evidence Code ontology
- Taxon lists must have Publication Taxon recorded for each row; it would be efficient to have Pub Taxon autofilled from Valid Taxon cell where they are the same name, and manually typed where Pub Taxon is a synonym.