Difference between revisions of "Conceptual Model of Data Warehouse"
m (Conceptual schema moved to Conceptual schema of the Phenoscape data warehouse: Misleading title) |
|||
| (3 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
[[Image:DataWarehouseLogicalModel.png]] | [[Image:DataWarehouseLogicalModel.png]] | ||
| + | |||
| + | ==Description of data model== | ||
| + | |||
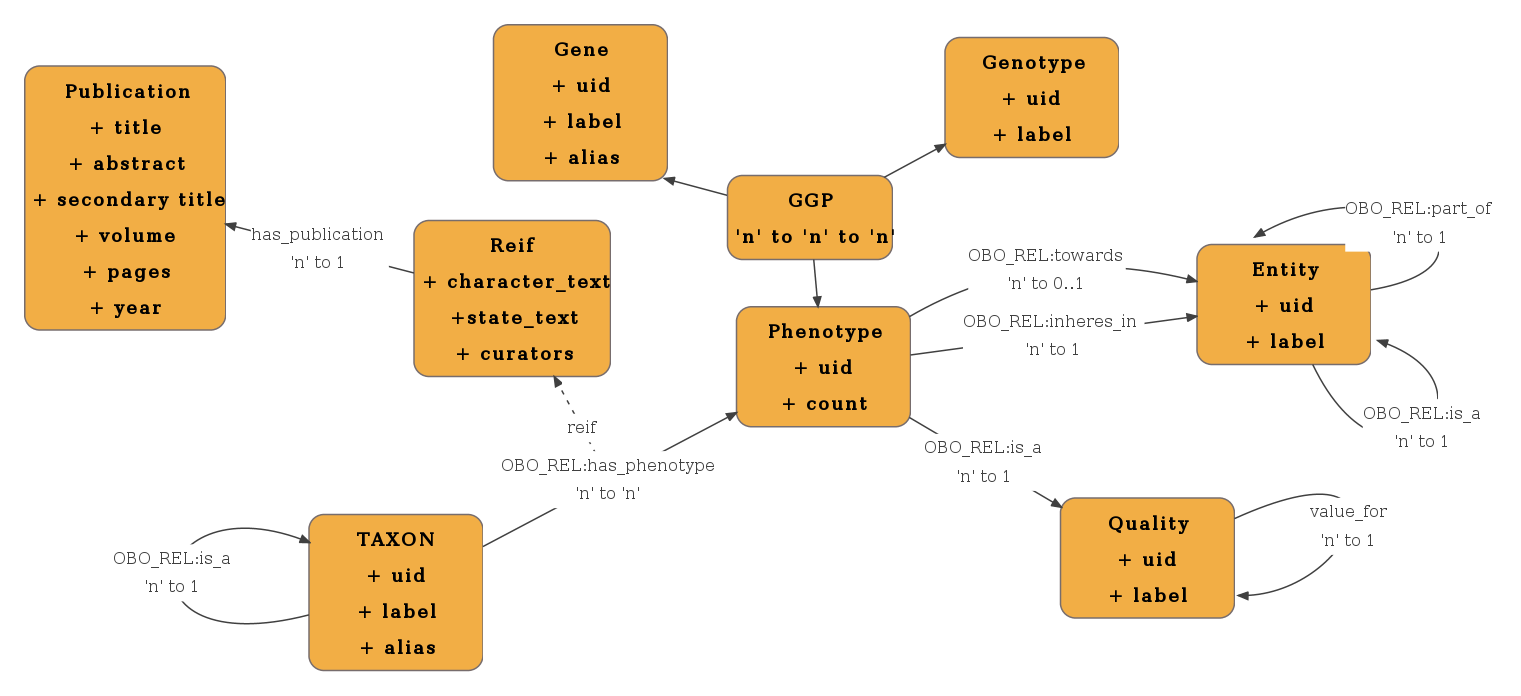
| + | The data warehouse has been designed with the intent of maximizing the efficiency of queries executed on the Phenoscape knowledge base. For phenotype queries, we need to know the phenotype in question, the taxa or genes which are associated with that phenotype, as well as the entity and quality associated with that phenotype. We also need to find the character, which the quality is associated with. For example if the quality is ''reduced number of'', the character in question would be ''count''. | ||
| + | |||
| + | To effectively execute this query, the phenotype centric model of the data warehouse is designed as follows (concepts and attributes are capitalized). A taxon or gene may be associated with one or more PHENOTYPE(s) and a PHENOTYPE may be associated with one or more genes or taxa. A PHENOTYPE is associated with exactly one ENTITY and one QUALITY. A QUALITY may be associated with one or more PHENOTYPE(s). Further, a QUALITY is associated with exactly one CHARACTER. A CHARACTER may be associated with one or more QUALITYs. The taxon or gene concepts have been merged into a SUBJECT concept. | ||
| + | |||
| + | For queries for provenance data about taxon to phenotype assertions, we need to find the publication the assertion is extracted from, the specific text from the publication about character and state, as well as the curators' comments about the assertion. | ||
| + | |||
| + | To effectively execute these 'metadata' queries, the provenance data is modeled as an association attribute. For every instance of the association between a SUBJECT and a PHENOYPE, we capture CHARACTER. STATE, CURATORS, and PUBLICATION. | ||
Latest revision as of 15:30, 8 September 2009
Description of data model
The data warehouse has been designed with the intent of maximizing the efficiency of queries executed on the Phenoscape knowledge base. For phenotype queries, we need to know the phenotype in question, the taxa or genes which are associated with that phenotype, as well as the entity and quality associated with that phenotype. We also need to find the character, which the quality is associated with. For example if the quality is reduced number of, the character in question would be count.
To effectively execute this query, the phenotype centric model of the data warehouse is designed as follows (concepts and attributes are capitalized). A taxon or gene may be associated with one or more PHENOTYPE(s) and a PHENOTYPE may be associated with one or more genes or taxa. A PHENOTYPE is associated with exactly one ENTITY and one QUALITY. A QUALITY may be associated with one or more PHENOTYPE(s). Further, a QUALITY is associated with exactly one CHARACTER. A CHARACTER may be associated with one or more QUALITYs. The taxon or gene concepts have been merged into a SUBJECT concept.
For queries for provenance data about taxon to phenotype assertions, we need to find the publication the assertion is extracted from, the specific text from the publication about character and state, as well as the curators' comments about the assertion.
To effectively execute these 'metadata' queries, the provenance data is modeled as an association attribute. For every instance of the association between a SUBJECT and a PHENOYPE, we capture CHARACTER. STATE, CURATORS, and PUBLICATION.