Difference between revisions of "Complex Query Specification"
(→Complex Query Specification) |
(→Complex Query Specification) |
||
| Line 3: | Line 3: | ||
== Version 4 == | == Version 4 == | ||
| + | [[Image:NewAdvancedQuery.001.png]] | ||
Revision as of 18:41, 11 March 2010
Contents
Complex Query Specification
Version 4
Notes for a Version 3
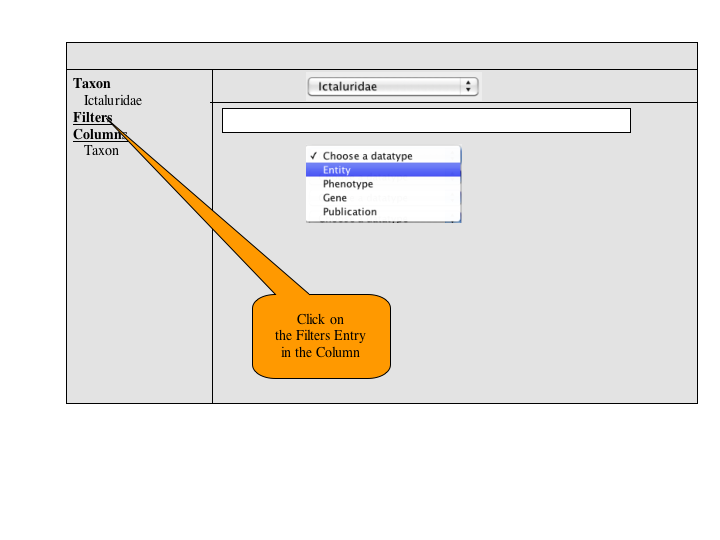
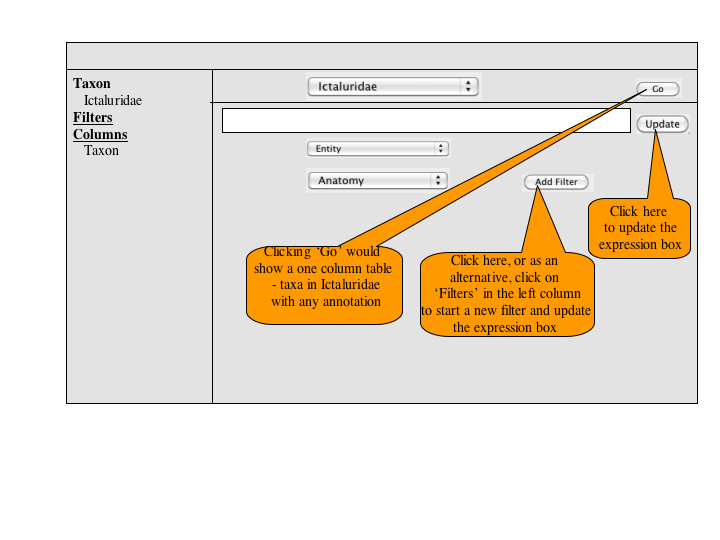
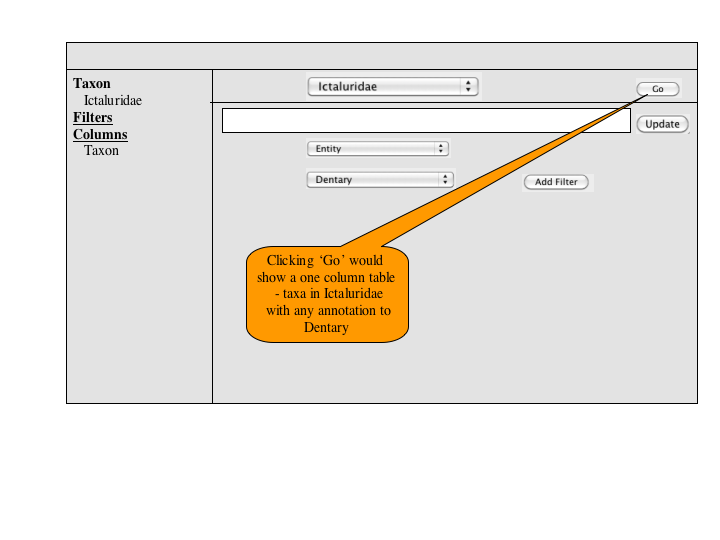
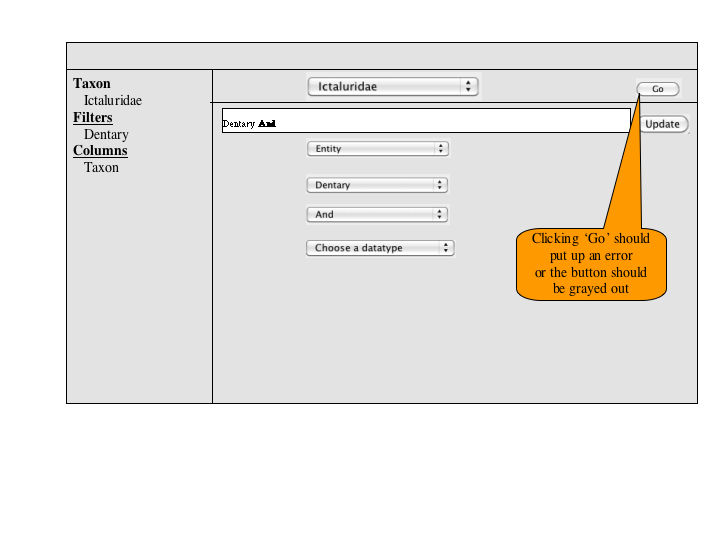
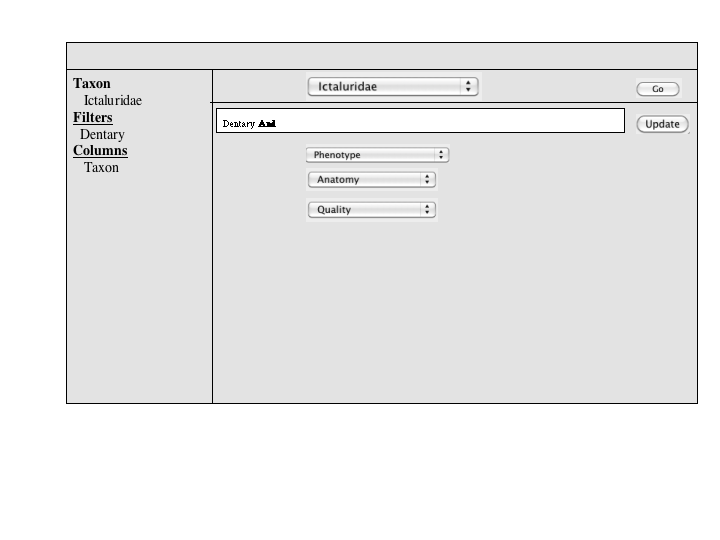
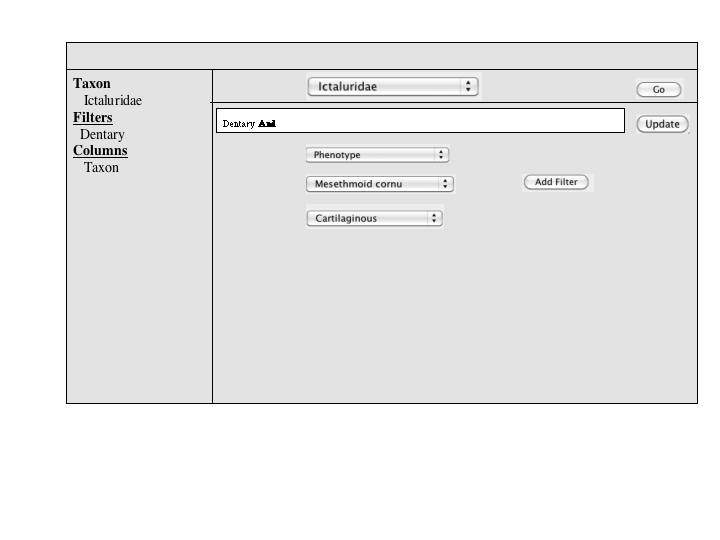
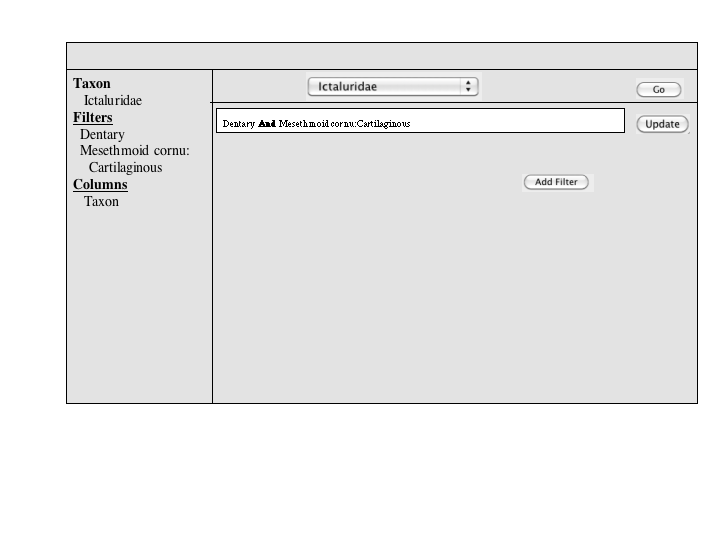
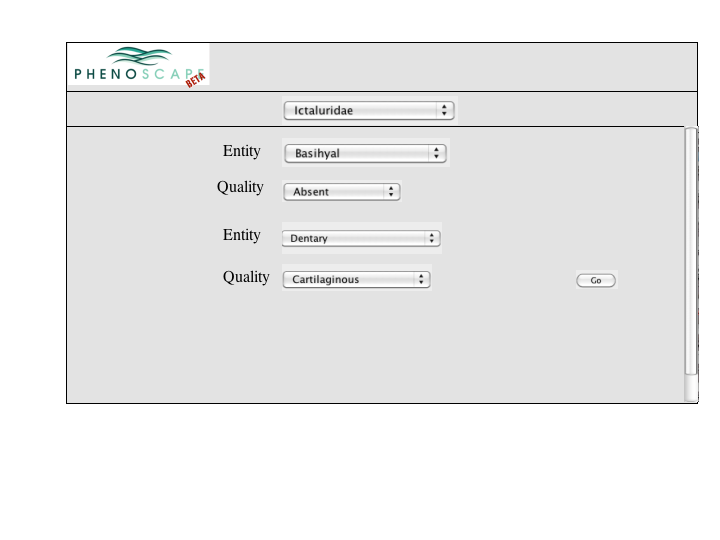
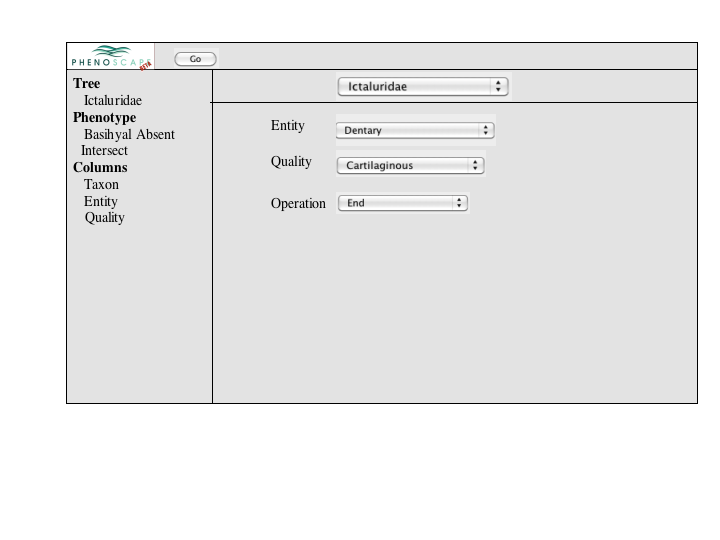
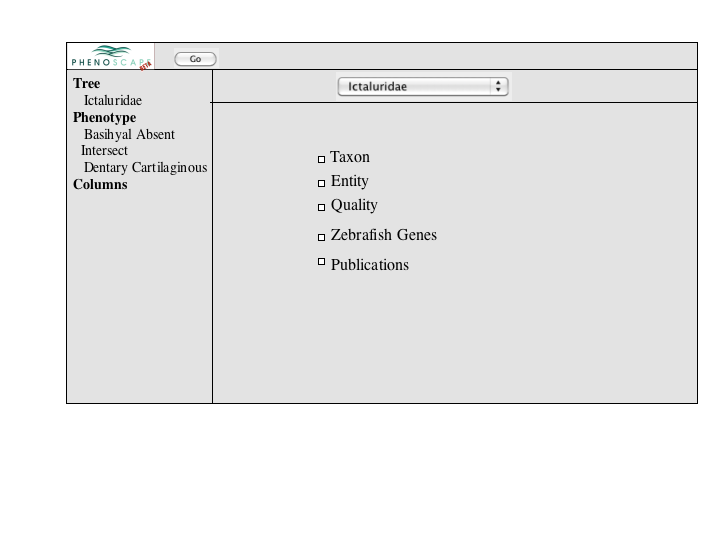
A number of approaches to a complex query have been suggested. Since Jim has added a simple query to many of the summary pages, the focus of a complex query is on covering the types of queries that can't be expressed in the standard query. An appropriate visualization of a query ought to be very explicit and could, secondarily let users check the semantics of a simple query (e.g., make the implicit disjunctions within categories and conjunctions between categories explicit). To this latter end, and as a way for users to discover how complex queries work, a button or link to a complex query page should be available at the bottom of every simple query panel. When selected, the contents of the simple query will be copied into the complex query page, establishing both a starting point for the user to build on and a correspondence back to the simple query.
List of suggested query models
- [Biomart]
- [Strategies Web Development Kit]
- examples at [PlasmoDB].
- [Phenomizer]
- [Ontogrator]
- Other 'list oriented' queries (e.g., fogbugz, mail.app, etc.)
Version 2
Entry from Taxon Page
Entry from Phenotype Page
Entry from Gene Page
Entry from Publication Page
[[Image:BioMart126_18.png]
Version 1
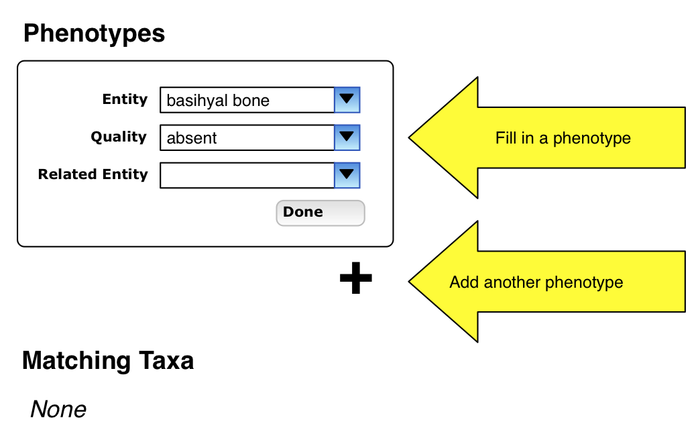
Entry of phenotype specification
Single Pane Set Algebra interface
Phenotype on Tree Mapping Specification
Biomart style interface
Version 0
- user enters a phenotype specification, presses Done