Difference between revisions of "Project Plan"
Paula Mabee (talk | contribs) (→Term broker (collab with NCBO)) |
Paula Mabee (talk | contribs) (→2. Ontology development and coordination (Wasila to fill in)) |
||
| Line 35: | Line 35: | ||
=== Target milestone: Year 4:July 1, 2015 === | === Target milestone: Year 4:July 1, 2015 === | ||
| − | =2. Ontology development and coordination (Wasila | + | =2. Ontology development and coordination (Wasila)= |
==Anatomy ontologies== | ==Anatomy ontologies== | ||
==Taxonomy ontologies== | ==Taxonomy ontologies== | ||
Revision as of 14:38, 10 October 2011
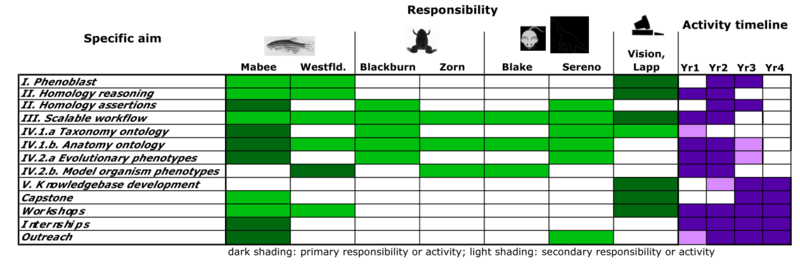
Much of the schedule below is based on the 1 July 2011 funded NSF grant. Our development schedule is revised as shown in the figure below in concert with budget cuts. Relative to the original proposal the support for curation, particularly in the model organism databases, is significantly reduced. This will impact our ability to respond as any problems arise with integration of the evolutionary data and somewhat compromise our ability to rigorously evaluate the tools being developed. Additionally, we will scale back the development goals for the semantic similarity search engine, focussing our efforts on achieving scalability and speedup. We have also frontloaded the plans for execution of the NLP work to achieve a scalable workflow as early as possible in the process.
Contents
- 1 1. Scalable workflow
- 1.1 Target milestone: First quarter: October 1, 2011
- 1.2 Target milestone: Second quarter: January 1, 2012
- 1.3 Target milestone: End of Year 1:July 1, 2012
- 1.4 Target milestone: Year 2:July 1, 2013
- 1.5 Target milestone: Year 3:July 1, 2014
- 1.6 Target milestone: Year 4:July 1, 2015
- 1.7 Term broker, in collaboration with NCBO (Hilmar)
- 2 2. Ontology development and coordination (Wasila)
- 3 3. Phenotype annotation
- 4 4. Homology (Paula to fill in and check with Hilmar)
- 5 5. Semantic similarity search engine (aka Phenoblast) and OBD/OWL (Todd, Jim? to fill in)
- 6 7. Capstone
1. Scalable workflow
Curation of legacy phenotypes from the literature is a major bottleneck. The overall objective of this part of our work is to improve the efficiency with which curators can find accurate terms, add missing terms, etc.
Primary personnel: Hong Cui, Jim, Todd, UNC postdoc, MS student
Target milestone: First quarter: October 1, 2011
Objective: Develop NLP to generate potential ontology terms and candidate EQ's in Phenex
- Generate list of terms to be added to ontologies (Hong)
- Evaluate accuracy of automated EQs on 50 character test set, refine testing set and methodology, begin development of Phenex ‘EQ suggestion’ interface requirements/specifications
- Identify corpus of publications, extract character descriptions
- Generate entities and qualities that can be identified, with frequency and ontology match
Action items:
- Wasila to send Hong the PDFs containing characters from the 50 char list - done
- Action item: Wasila to send Paul a curated pub
- Action Item: Jim will send Hong full database report with character text and EQ assignments
- Jim: to enumerate pros and cons for MX (web-based) and Phenex for first phone call, estimate development time
- Hong: Hire MS student at AZ
- Todd: Hire UNC postdoc
Target milestone: Second quarter: January 1, 2012
Target milestone: End of Year 1:July 1, 2012
Target milestone: Year 2:July 1, 2013
Target milestone: Year 3:July 1, 2014
Target milestone: Year 4:July 1, 2015
Term broker, in collaboration with NCBO (Hilmar)
Target milestone: First quarter: October 1, 2011
Target milestone: Second quarter: January 1, 2012
Target milestone: End of Year 1:July 1, 2012
Target milestone: Year 2:July 1, 2013
Target milestone: Year 3:July 1, 2014
Target milestone: Year 4:July 1, 2015
2. Ontology development and coordination (Wasila)
Anatomy ontologies
Taxonomy ontologies
3. Phenotype annotation
Evolutionary phenotypes (Paula)
The objective is to transform the characters and character states from published phylogenetic studies into ontology-based descriptions ('Evolutionary phenotypes'), with a focus on fin and limb morphology. This will require the development of a list of papers to be curated, re-evaluation of software curation tool, training of personnel in use of curation software and ontology development, and development of appropriate ontologies.
Personnel: Paula*, David, Paul, Wasila, Jim, and postdoctoral fellows
- Coordinator
Target milestone: First quarter: October 1, 2011
Objective: Develop a prioritized list of phylogenetic papers containing vertebrate fin/limb data for curation; evaluate curation tool; prepare KB for vertebrate data
Action items:
- Develop a list of priority papers (pdfs) to be curated. --Pmabee@usd.edu 10:37, 10 October 2011 (EDT)done except archosaurs
- Annotation for AmAO to begin after AmAO developed (August 2011)
- Training (ontology editor; annotation tool) for Paul & Nizar at NESCent
- Paul and Nizar will document in lists any additional terms and definitions before meeting; prepare to add them to AmAO at NESCent.
- Evaluate curation tool - do we need a new one?
- Jim will develop KB instance in parallel for Vertebrates (a new beta) this summer
Target milestone: Second quarter: January 1, 2012
Target milestone: End of Year 1:July 1, 2012
Target milestone: Year 2: July 1, 2013
- Annotation for AAO to begin in Year 2 (after cloning XAO)
Target milestone: Year 3: July 1, 2014
Target milestone: Year 4: July 1, 2015
Model organism phenotypes (Monte)
Objective: To annotate the skeletal phenotypes for fin and limb for genetic mutants of zebrafish, Xenopus, and mouse. The model organism (MOD) curators will initially prioritize comprehensive annotation of skeletal phenotypes for the fin and limb, and subsequently of skeletal phenotypes in general.
Involved personnel: ZFIN (Monte*, Ceri,Yvonne), Xenbase (Aaron, Christina), MGI (Judy, Terry)
- coordinator
Target milestone: First quarter: October 1, 2011
- Curation of expression and phenotypes
- Investigate additional funding through NSF (Todd) and NIH (Monte, Judy)
- Determine whether the current MP->EQ mapping is sufficient (i.e., the mapping that had been done with
George)? In particular, is the limb and limb girdle mapping complete?
Action items:
- Judy will check with Martin to see if limb mapping has done
- Determine who is responsible for completing the mapping and keeping it up to date? Cindy? How to coordinate with PATO?
- Are there outstanding problems with developmental phenotypes for mouse, i.e. how to incorporate the abstract mouse?
- Determine timeline for:
- Developing pipelines for uploading Xenbase and MGI phenotype data to Phenoscape
- Incorporating expression data into Phenoscape KB
Target milestone: Second quarter: January 1, 2012
Target milestone: End of Year 1:July 1, 2012
Target milestone: Year 2: July 1, 2013
- In year 2, Xenbase will begin curating phenotypes
Target milestone: Year 3: July 1, 2014
Target milestone: Year 4: July 1, 2015
4. Homology (Paula to fill in and check with Hilmar)
Homology reasoning
Homology assertions
5. Semantic similarity search engine (aka Phenoblast) and OBD/OWL (Todd, Jim? to fill in)
=6. Knowledgebase reasoning and development
7. Capstone
component 1
DBI 1062404 and 1062542: Collaborative research: ABI Development: Ontology-enabled reasoning across phenotypes from evolution and model organisms 1. Technical description of the project. An award is made to the University of South Dakota and the University of North Carolina to develop ontology-driven tools for machine reasoning over large volumes of phenotype data. A fast semantic similarity engine will be developed to allow searches for evolutionary transitions and mutant genes characterized by similar phenotypic profiles. An ontological framework for reasoning over homology will be developed to allow rigorous reasoning over evolutionary diverse lineages. Natural language processing tools will be developed to improve upon the efficiency of mining phenotype data from the literature and improving data consistency. This suite of tools will be tested on a large number of skeletal phenotypes from diverse fossil and modern vertebrates. Taxonomic and anatomical ontologies for vertebrates will be augmented and hypotheses of anatomical homology formally encoded. The ontologies and software tools, together with phenotypes extracted from the vertebrate systematic literature, will be integrated in the knowledgebase with genetic and phenotype data from three vertebrate model organisms: zebrafish (Danio rerio), African clawed frog (Xenopus laevis), and mouse (Mus musculus). The knowledgebase will be exposed to generic reasoners using semantic web standards. The system will be validated by its success in retrieving candidate genes for the well-studied vertebrate fin-limb transition and other major events in skeletal evolution.
2. Non-technical explanation of the project's broader significance and importance. Human-readable descriptions of “phenotypic” properties such as anatomy and behavior are not well-suited to computational analysis. Yet, in evolutionary biology, genetics and development, computational assistance is necessary to discover patterns within the enormous volumes of descriptive phenotype data that are being reported in the literature and in online databases. Ontologies are structured, controlled vocabularies that can be applied to collections of descriptive data to permit logical reasoning to be used. Using the evolutionary transition from fins to limbs as a test system, this project will develop ontologically-aware software that allows users to discover similar sets of phenotypes for different taxa or mutant genes within large and diverse datasets. The evolutionary breadth of the test data requires the development of a rigorous framework for reasoning over hypotheses of homology. Another goal is to develop and evaluate natural language processing tools for efficiently capturing ontological descriptions of phenotype from the descriptions available in the published literature. Phenotype data from the systematic literature for both extinct and extant vertebrates will be combined with mutant phenotype data from three vertebrate genetic models: zebrafish (Danio rerio), frog (Xenopus laevis), and mouse (Mus musculus). The suite of tools will be validated by recovering developmental genetic pathways that underlie the evolutionary transition from fin to limb in vertebrates, and refined by iterative testing with domain bioinformaticians on the project and biologists from the broader user community.
3. Indicate how your project addresses criteria specific to Development A broad community of users will participate through the lifecycle of this project in the development of community standards and resources for the interoperability and computability of phenotypic knowledge. This will be achieved through workshops, usability testing sessions, and coordination with key research networks. Stakeholder ownership will be enhanced by rapid and open release of a variety of products that we anticipate to be of immediate and enduring value to the greater biology community, including tools for streamlining data curation and performing large-scale semantic similarity searches, high quality vertebrate taxonomy and anatomy ontologies, and standards for reasoning over homology. We will provide a unique training environment for students, postdocs and summer interns, including Native Americans through outreach at the University of South Dakota and minority and female students though a collaboration with Project Exploration at the University of Chicago. Project progress and outcomes will be disseminated through both traditional and online outlets for scholarly communication (including blog posts at mailing lists); the primary web presence will be at https://www.phenoscape.org/wiki/.