Difference between revisions of "KB build process"
Jim Balhoff (talk | contribs) (→OWL conversion) |
Jim Balhoff (talk | contribs) (→RDF data model) |
||
| (44 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
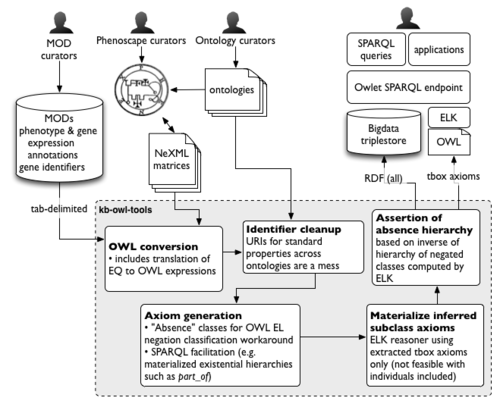
| − | + | The Phenoscape KB build process goes through several steps in converting input data sources to a queryable knowledgebase. This page provides some description for each of the steps, most or all of which are implemented in the [https://github.com/phenoscape/phenoscape-owl-tools phenoscape-owl-tools] project. The result of this pipeline is a unified OWL dataset containing the ontologies and data. The result is loaded as RDF into a [[Bigdata|Blazegraph]] triplestore. | |
| − | + | [[Image:PhenoscapeKB-build-process.png|right|500px]] | |
==OWL conversion== | ==OWL conversion== | ||
The Phenoscape Knowledgebase works as a single unified OWL model. While some inputs (e.g. the shared ontologies such as [http://uberon.org/ Uberon] and [http://purl.obolibrary.org/obo/pato PATO]) are natively distributed as OWL documents, others are converted to OWL from some other representation. In doing so the inputs are, as far as possible, converted to a shared data model. EQ annotations are converted to a specific semantic representation. | The Phenoscape Knowledgebase works as a single unified OWL model. While some inputs (e.g. the shared ontologies such as [http://uberon.org/ Uberon] and [http://purl.obolibrary.org/obo/pato PATO]) are natively distributed as OWL documents, others are converted to OWL from some other representation. In doing so the inputs are, as far as possible, converted to a shared data model. EQ annotations are converted to a specific semantic representation. | ||
===Examples=== | ===Examples=== | ||
| − | * ZFIN | + | * ZFIN phenotype annotations are provided at [http://zfin.org/downloads their data download site]. We translate each row into a phenotype instance: an individual instance of some PATO quality. Here are the column headers from the ZFIN website, followed by representative OWL syntax (referencing column numbers where actual URI values would be, and somewhat casual with other identifiers): |
| + | |||
<table class="summary1"> | <table class="summary1"> | ||
| − | + | <tr> | |
| − | + | <td style="font-weight: bold; background-color: #CCFFFF; text-align: center">1</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF; text-align: center">2</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF; text-align: center">3</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF; text-align: center">4</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF; text-align: center">5</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF; text-align: center">6</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF; text-align: center">7</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF; text-align: center">8</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF; text-align: center">9</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF; text-align: center">10</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF; text-align: center">11</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF; text-align: center">12</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF; text-align: center">13</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF; text-align: center">14</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF; text-align: center">15</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF; text-align: center">16</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF; text-align: center">17</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF; text-align: center">18</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF; text-align: center">19</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF; text-align: center">20</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF; text-align: center">21</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF; text-align: center">22</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF; text-align: center">23</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF; text-align: center">24</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF; text-align: center">25</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF; text-align: center">26</td> | |
| − | + | ||
| − | + | </tr> | |
| − | + | <tr> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF;">ID</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF;">Gene Symbol</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF;">Gene ID</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF;">Affected Structure or Process 1 subterm ID</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF;">Affected Structure or Process 1 subterm Name</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF;">Post-composed Relationship ID</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF;">Post-composed Relationship Name</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF;">Affected Structure or Process 1 superterm ID</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF;">Affected Structure or Process 1 superterm Name</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF;">Phenotype Keyword ID</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF;">Phenotype Keyword Name</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF;">Phenotype Tag</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF;">Affected Structure or Process 2 subterm ID</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF;">Affected Structure or Process 2 subterm name</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF;">Post-composed Relationship (rel) ID</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF;">Post-composed Relationship (rel) Name</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF;">Affected Structure or Process 2 superterm ID</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF;">Affected Structure or Process 2 superterm name</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF;">Genotype ID</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF;">Genotype Display Name</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF;">Knockdown Reagent ID</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF;">Start Stage ID</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF;">End Stage ID</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF;">Genotype Environment ID</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF;">Publication ID</td> | |
| − | + | ||
| − | + | <td style="font-weight: bold; background-color: #CCFFFF;">Figure ID</td> | |
| − | + | ||
| − | + | </tr> | |
| − | + | </table> | |
| − | + | ||
| + | <pre> | ||
| + | Individual: <uuid_1> | ||
| + | Types: ps:AnnotatedPhenotype, | ||
| + | <uuid_2> | ||
| + | Facts: ps:associated_with_gene <col_3>, | ||
| + | ps:associated_with_taxon NCBITaxon:zebrafish, | ||
| + | dc:source <col_26> | ||
| + | |||
| + | Class: <uuid_2> | ||
| + | SubClassOf: <col_10> and | ||
| + | (inheres_in some (<col_4> and (<col_6> some <col_8>))) and | ||
| + | (towards some (<col_13> and (<col_15> some <col_17>))) | ||
| + | </pre> | ||
| + | |||
| + | * Annotations from other MODs are similarly structured; however some such as MGI use precoordinated phenotype classes instead of an EQ template. In that case, the phenotype instance is simply an instance of the stated [http://www.informatics.jax.org/searches/MP_form.shtml MP] phenotype term. | ||
==Identifier cleanup== | ==Identifier cleanup== | ||
| − | Several | + | Several common OWL properties (''part_of'', ''has_part'', ''develops_from'', etc.) are conceptually shared across ontology and annotation resources, facilitating data integration. However, unlike class identifiers, identifiers for properties are often not standardized and they may not properly reference shared terms (usually because of poor tool support rather than user intent). We maintain a [https://github.com/phenoscape/phenoscape-owl-tools/blob/master/src/main/scala/org/phenoscape/owl/PropertyNormalizer.scala table of "alternative" URIs] for common properties as we observe them in our data inputs. We could create equivalence axioms between these, but instead we just standardize all incoming content. This saves the reasoner some work and also makes it much easier to query across data using standard URIs, especially when not using a reasoner. |
==Axiom generation== | ==Axiom generation== | ||
| − | + | ==="Absence" classes for OWL EL negation classification workaround=== | |
| − | + | We model absence phenotypes as an instance of the quality [http://purl.obolibrary.org/obo/PATO_0002000 lacks all parts of type] which ''inheres_in'' the [http://purl.obolibrary.org/obo/UBERON_0000468 whole organism] and is ''towards'' a type, the class of structure that is absent, i.e. <code>''towards'' value '''X'''</code>. This approach allows us to have a phenotype class for the condition, but directly using a class as a property value in this way is outside of DL reasoning. So to classify our absence phenotypes, we also add an equivalence to <code>not (''has_part'' some '''X''')</code>. ''TODO—this should probably be'' <code>inheres_in some (not (''has_part'' some '''X'''))</code>. Because we are confined to using the ELK reasoner (for scalability reasons) class expressions using <code>not</code>, which is excluded from OWL EL, will still not be classified. For this reason we must pregenerate an "absence" phenotype for each known anatomical structure. These generated classes are named in a standardized way so that they can be arranged into a hierarchy by the "Negation Hierarchy Asserter" ([[KB_build_process#Assertion_of_absence_hierarchy|below]]). | |
| − | + | ||
| + | For more on modeling of absence phenotypes, see [[Absence Phenotypes in OWL]]. | ||
| + | |||
| + | ===SPARQL facilitation (e.g. materialized existential hierarchies such as part_of)=== | ||
| + | ==="EQ character" classes: combinations of anatomical entities and PATO attribute terms=== | ||
| + | For the purposes of the EQ character matrix project, a set of phenotypic trait classes is generated using every combination of anatomical entity, from Uberon, and the small set of "attribute" qualities, from PATO. Each combination is composed into a class with semantics <code>'''attribute''' and (''inheres_in'' some '''entity''') and '''EQCharacterToken'''</code>. When phenotype annotations from NeXML files are translated into OWL classes, each of these OWL classes is asserted to be a subclass of <code>'''EQCharacterToken'''</code>. The token class ensures that every annotation phenotype class will be classified by the reasoner as a direct subclass of one of these phenotypic trait classes. | ||
==Materialization of inferred axioms== | ==Materialization of inferred axioms== | ||
| − | ELK reasoner | + | The ELK reasoner is used to compute the inferred class hierarchy. The reasoner is provided only with tbox axioms (statements about classes, not properties of individuals), which greatly improves reasoning speed. However this limitation must be kept in mind when considering data modeling and intended querying capabilities. |
==Assertion of absence hierarchy== | ==Assertion of absence hierarchy== | ||
| − | + | The [https://github.com/phenoscape/phenoscape-owl-tools/blob/master/src/main/scala/org/phenoscape/owl/NegationHierarchyAsserter.scala Negation Hierarchy Asserter] looks for classes which are linked via a special annotation property, "negates", to some other named class. For each class N which negates a class C, any classes which negate superclasses of C are asserted as subclasses of N. Any classes which negate subclasses of C are asserted as superclasses of N. This allows us to compute a hierarchy for a specific set of negations (such as the set of "absence" classes) using the hierarchy of their complements. | |
| + | |||
| + | ==RDF data model== | ||
| + | The RDF data model resulting from the KB build process is depicted in this graph diagram. This is the form in which the data are loaded into Blazegraph, and subsequently queried using SPARQL to drive KB web services. | ||
| + | |||
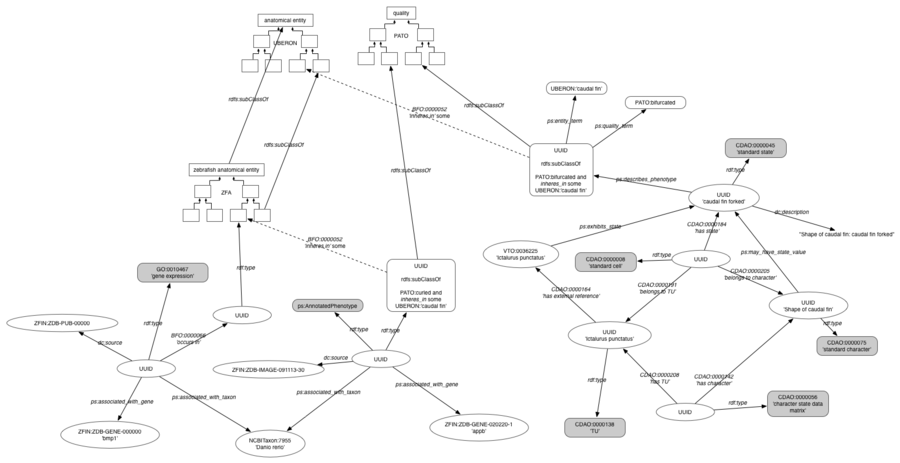
| + | [[Image:PhenoscapeKB-RDF-model.png|900px]] | ||
[[Category:Reasoning]] | [[Category:Reasoning]] | ||
Latest revision as of 18:14, 27 July 2015
The Phenoscape KB build process goes through several steps in converting input data sources to a queryable knowledgebase. This page provides some description for each of the steps, most or all of which are implemented in the phenoscape-owl-tools project. The result of this pipeline is a unified OWL dataset containing the ontologies and data. The result is loaded as RDF into a Blazegraph triplestore.
Contents
OWL conversion
The Phenoscape Knowledgebase works as a single unified OWL model. While some inputs (e.g. the shared ontologies such as Uberon and PATO) are natively distributed as OWL documents, others are converted to OWL from some other representation. In doing so the inputs are, as far as possible, converted to a shared data model. EQ annotations are converted to a specific semantic representation.
Examples
- ZFIN phenotype annotations are provided at their data download site. We translate each row into a phenotype instance: an individual instance of some PATO quality. Here are the column headers from the ZFIN website, followed by representative OWL syntax (referencing column numbers where actual URI values would be, and somewhat casual with other identifiers):
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 |
| ID | Gene Symbol | Gene ID | Affected Structure or Process 1 subterm ID | Affected Structure or Process 1 subterm Name | Post-composed Relationship ID | Post-composed Relationship Name | Affected Structure or Process 1 superterm ID | Affected Structure or Process 1 superterm Name | Phenotype Keyword ID | Phenotype Keyword Name | Phenotype Tag | Affected Structure or Process 2 subterm ID | Affected Structure or Process 2 subterm name | Post-composed Relationship (rel) ID | Post-composed Relationship (rel) Name | Affected Structure or Process 2 superterm ID | Affected Structure or Process 2 superterm name | Genotype ID | Genotype Display Name | Knockdown Reagent ID | Start Stage ID | End Stage ID | Genotype Environment ID | Publication ID | Figure ID |
Individual: <uuid_1> Types: ps:AnnotatedPhenotype, <uuid_2> Facts: ps:associated_with_gene <col_3>, ps:associated_with_taxon NCBITaxon:zebrafish, dc:source <col_26> Class: <uuid_2> SubClassOf: <col_10> and (inheres_in some (<col_4> and (<col_6> some <col_8>))) and (towards some (<col_13> and (<col_15> some <col_17>)))
- Annotations from other MODs are similarly structured; however some such as MGI use precoordinated phenotype classes instead of an EQ template. In that case, the phenotype instance is simply an instance of the stated MP phenotype term.
Identifier cleanup
Several common OWL properties (part_of, has_part, develops_from, etc.) are conceptually shared across ontology and annotation resources, facilitating data integration. However, unlike class identifiers, identifiers for properties are often not standardized and they may not properly reference shared terms (usually because of poor tool support rather than user intent). We maintain a table of "alternative" URIs for common properties as we observe them in our data inputs. We could create equivalence axioms between these, but instead we just standardize all incoming content. This saves the reasoner some work and also makes it much easier to query across data using standard URIs, especially when not using a reasoner.
Axiom generation
"Absence" classes for OWL EL negation classification workaround
We model absence phenotypes as an instance of the quality lacks all parts of type which inheres_in the whole organism and is towards a type, the class of structure that is absent, i.e. towards value X. This approach allows us to have a phenotype class for the condition, but directly using a class as a property value in this way is outside of DL reasoning. So to classify our absence phenotypes, we also add an equivalence to not (has_part some X). TODO—this should probably be inheres_in some (not (has_part some X)). Because we are confined to using the ELK reasoner (for scalability reasons) class expressions using not, which is excluded from OWL EL, will still not be classified. For this reason we must pregenerate an "absence" phenotype for each known anatomical structure. These generated classes are named in a standardized way so that they can be arranged into a hierarchy by the "Negation Hierarchy Asserter" (below).
For more on modeling of absence phenotypes, see Absence Phenotypes in OWL.
SPARQL facilitation (e.g. materialized existential hierarchies such as part_of)
"EQ character" classes: combinations of anatomical entities and PATO attribute terms
For the purposes of the EQ character matrix project, a set of phenotypic trait classes is generated using every combination of anatomical entity, from Uberon, and the small set of "attribute" qualities, from PATO. Each combination is composed into a class with semantics attribute and (inheres_in some entity) and EQCharacterToken. When phenotype annotations from NeXML files are translated into OWL classes, each of these OWL classes is asserted to be a subclass of EQCharacterToken. The token class ensures that every annotation phenotype class will be classified by the reasoner as a direct subclass of one of these phenotypic trait classes.
Materialization of inferred axioms
The ELK reasoner is used to compute the inferred class hierarchy. The reasoner is provided only with tbox axioms (statements about classes, not properties of individuals), which greatly improves reasoning speed. However this limitation must be kept in mind when considering data modeling and intended querying capabilities.
Assertion of absence hierarchy
The Negation Hierarchy Asserter looks for classes which are linked via a special annotation property, "negates", to some other named class. For each class N which negates a class C, any classes which negate superclasses of C are asserted as subclasses of N. Any classes which negate subclasses of C are asserted as superclasses of N. This allows us to compute a hierarchy for a specific set of negations (such as the set of "absence" classes) using the hierarchy of their complements.
RDF data model
The RDF data model resulting from the KB build process is depicted in this graph diagram. This is the form in which the data are loaded into Blazegraph, and subsequently queried using SPARQL to drive KB web services.