EQ for character matrices
This document is under construction... --Jpb15 12:13, 21 May 2007 (EDT)
Encoding evolutionary character matrices using the EQ format (PATO formalism) currently presents some problems that may need to be resolved. I will try to describe the issues here.
The EQ format provides a "phenotype statement" documenting the phenotype of an individual organism (usually a genetic mutant). The anatomical structure being described is represented by the Entity term chosen from an anatomical ontology, and the aspect of that structure being described is the Quality term, chosen from the PATO ontology. These phenotype statements usually describe the value the mutant exhibits:
E="dorsal fin" Q="round" --> This fish has a rounded dorsal fin.
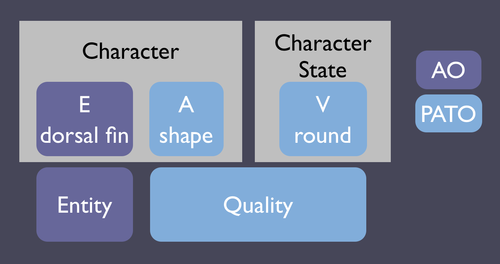
Evolutionary phenotype descriptions are often formatted as a character matrix. For a given set of species, a list of distinguishing characters is formulated, and species-specific value for each character is entered into the matrix. In this situation, each character (column in the matrix) represents an entity and attribute, and the character state cells contain values. Here is a graphical depiction of the relationship between evolutionary characters and the components of the EQ system:
The character (a column header in the matrix) is composed of an Entity and a Quality representing an attribute (e.g. "shape"). Values for this character are entered into the cells (e.g. "round). So you can see that the Q of EQ is represented in both the character and the character state. When EQ is used to describe mutant phenotypes, typically only the value is stated, since one can traverse back through the PATO hierarchy to find ancestor terms representing attributes - "round" is a child of "shape".
Clearly, evolutionary characters and character states can be represented using an EQ system as mutant phenotypes are. However there is a major difference in the two data models: the data formats being developed for mutant phenotype EQ statements store only a list of phenotype value statements, with no place to reference an independent character. So you may have a data set like this:
| Genotype | Entity | Quality |
|---|---|---|
| fish1 | dorsal fin | round |
| fish2 | dorsal fin | lobate |
| fish2 | dorsal fin | red |
| fish1 | pectoral fin | blue |
If these fish are different species, you can see that there are 2 values for the character "dorsal fin shape", 1 value for "dorsal fin color", and 1 value for "pectoral fin color". But nowhere in the existing EQ data formats is the character stored. We had to infer it from the PATO hierarchy to generate a matrix like this:
| Species | dorsal fin shape | dorsal fin color | pectoral fin color |
|---|---|---|---|
| fish1 | round | ? | blue |
| fish2 | lobate | red | ? |