Knowledgebase mockups
Contents
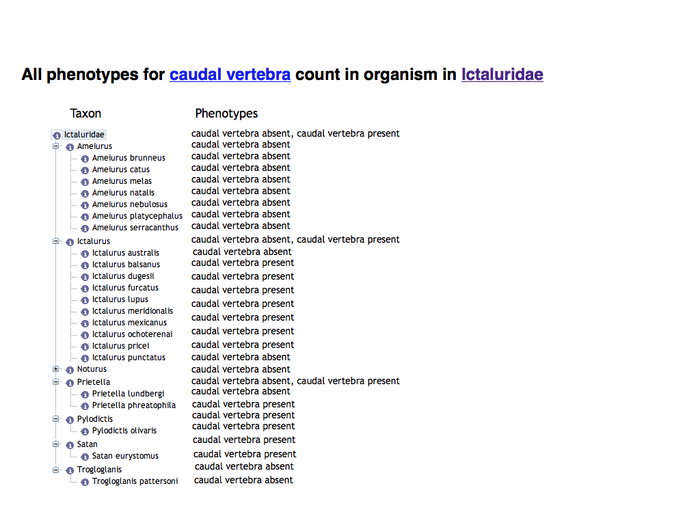
Grouping data by using an ontology tree view
Phenotype results
This would be an enhancement to the results currently displayed on a page like http://kb.phenoscape.org/phenotype/evo/TTO:10930/TAO:0000326/PATO:0000070/
The nearest common ancestor term of the annotated taxon terms is used as the root of the tree display. The only leaf terms displayed are those with annotations. Internal nodes show the union of the annotations to their descendant nodes.
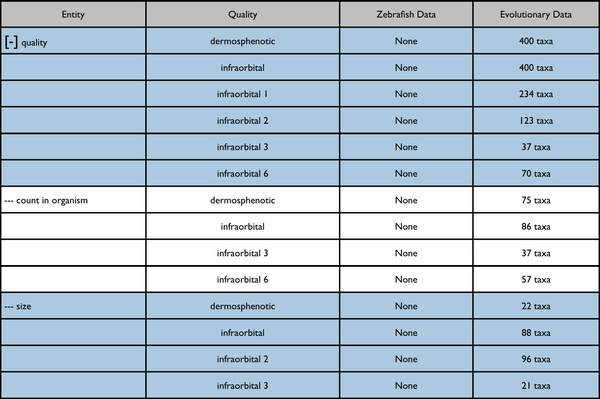
Search summary results
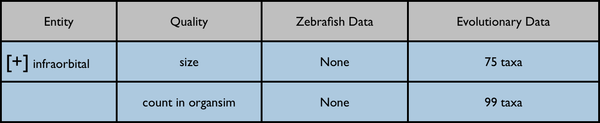
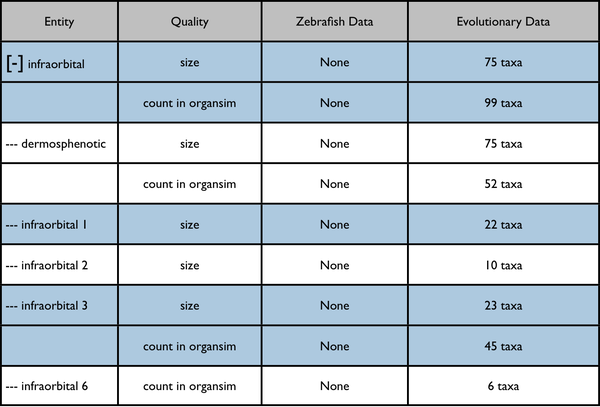
This would be an enhancement to the Phenotypes results currently displayed on a page like http://kb.phenoscape.org/search/anatomy/TAO:0000376
The leftmost column is intended to represent a disclosable tree display. The nearest common ancestor of the represented annotated anatomy terms is used as the root of the tree display. Each anatomy term has a flat list of the qualities annotated for it. Don't pay too much attention to the dummy numbers given in these results.
Root anatomy term "infraorbital" collapsed:
Root anatomy term "infraorbital" expanded:
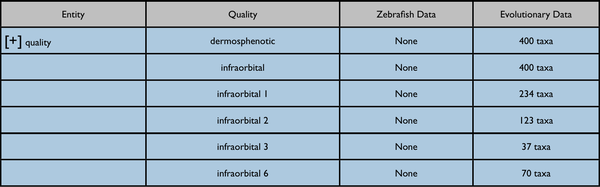
Possible further enhancement to page capabilities - redisplay data on a quality tree. The included results would be somewhat different.
Nearest quality ancestor collapsed:
Quality ancestor expanded:
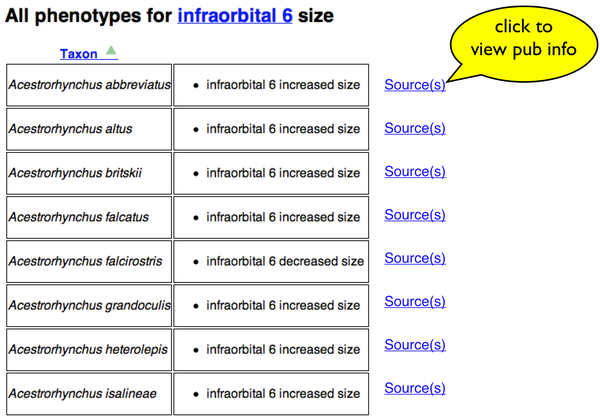
Display of citation and free-text basis for annotations
Here is a list of specific phenotype annotations resulting from a search. There is an added "Source(s)" link next to each one.
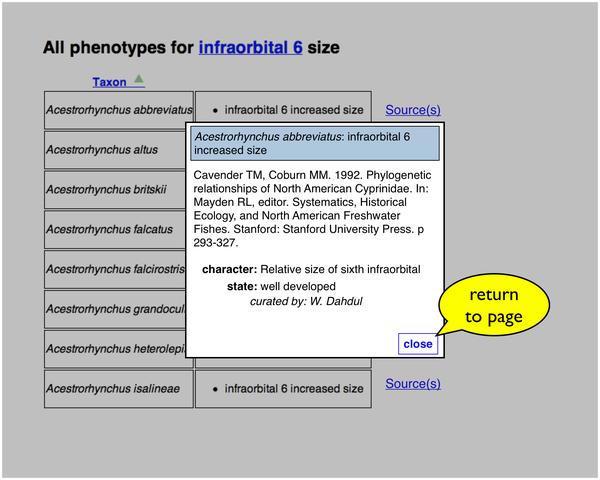
If the user clicks a "Source(s)" link, the citation information for a particular taxon-phenotype annotation is retrieved and displayed in a page overlay.
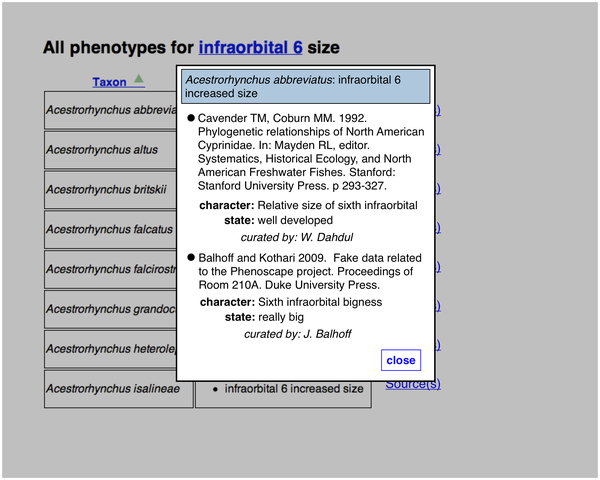
A particular taxon-phenotype annotation may have been asserted from more than one publication: