Transitive homology
This page explores the transitive closure of the homologous_to relation. Currently in OBD this is a binary class-level relation that holds between two taxon-specific anatomical entities.
I propose the following expansion to OWL-DL instance-level quantified axioms:
homologous_to(A,B) ==> Class: A SubClassOf: descendedFrom some (hasDescendant some B) Class: B SubClassOf: descendedFrom some (hasDescendant some A)
If we make the simplifying assumption that directlyDescendsFrom is functional then it follows that class-level homology is transitive. What are the consequences of making this strong assumption? We can explore this with existing phenoscape data.
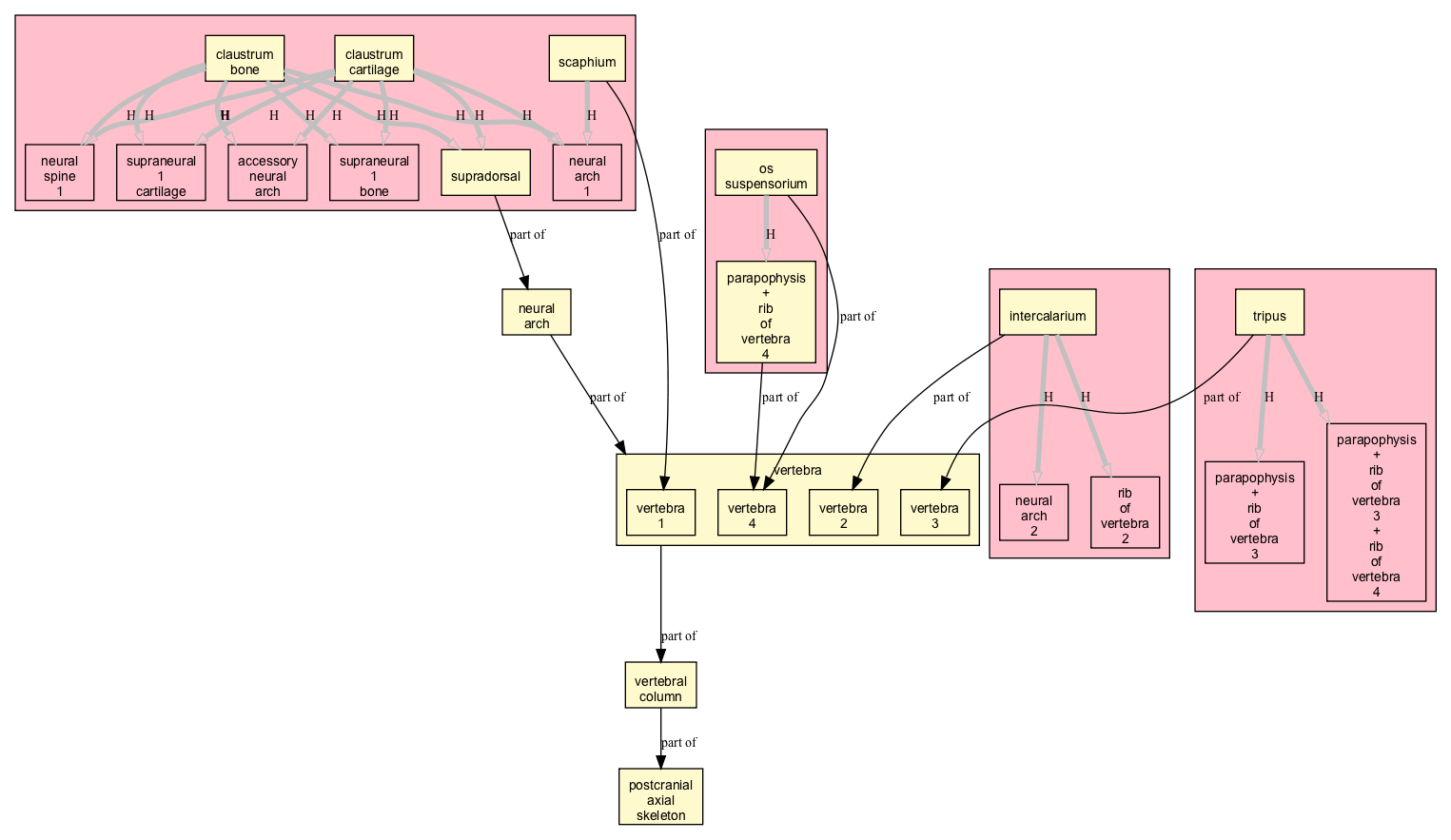
High Level Overview
This is a high level part_of view of all homology relationships, projected onto TAO:
- homology statements are generalized to the TAO, taxa not shown
- clusters of homology relationships are shown in 4 distinct boxes
- vertebra clustered into a box
- only relations shown are homology and part_of
Here we can see there are 4 distinct clusters, according to the vertebra number affected
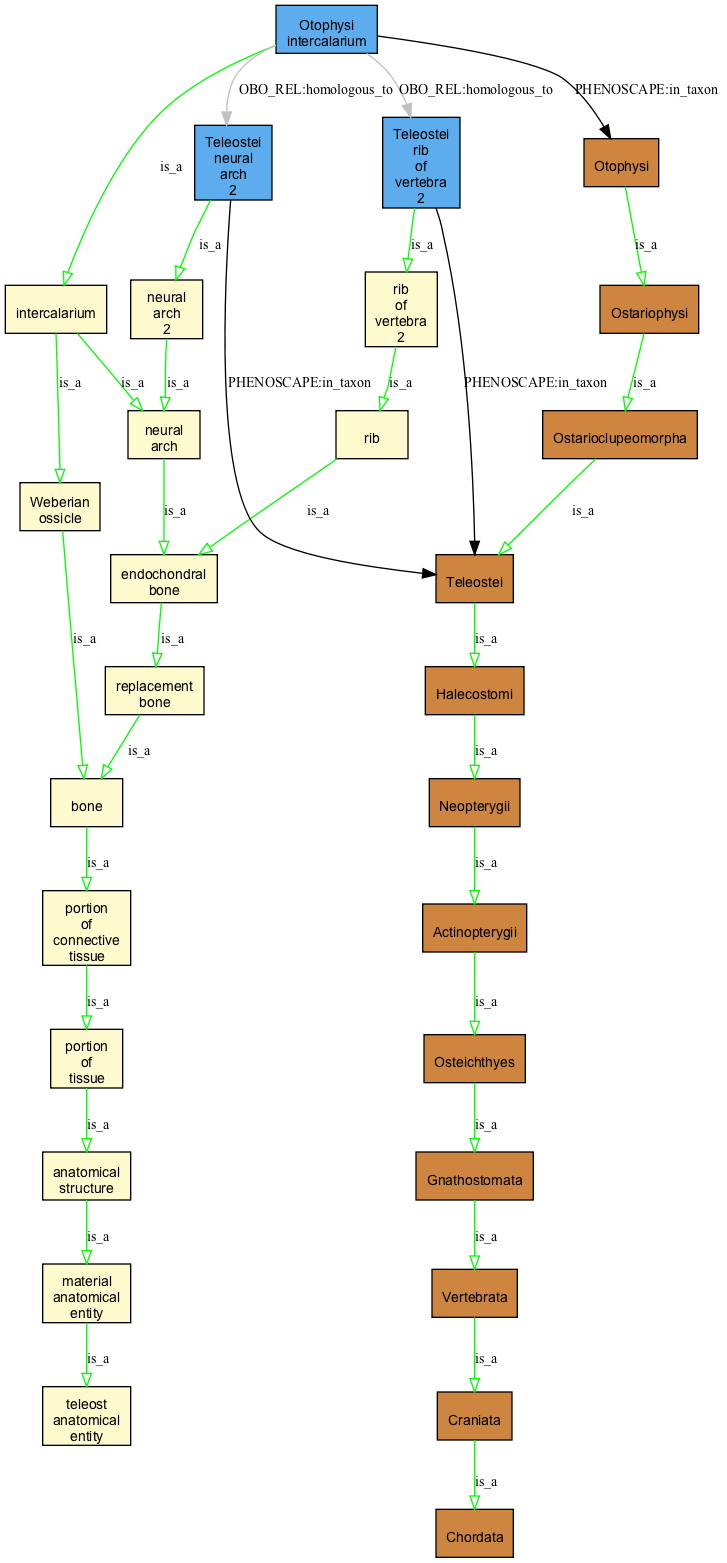
Otophysi intercalarium
this is annotated to be homologous to the neural arch 2 and vertebra 2 in Teleosti. From this, what can be said of the relationship between these two entities?
Note that for compactness reasons this diagram doesn't show the part_of relations.
- intercalarium part_of v2
presumably there should be some kind of relationship between "rib of v2" and "v2"
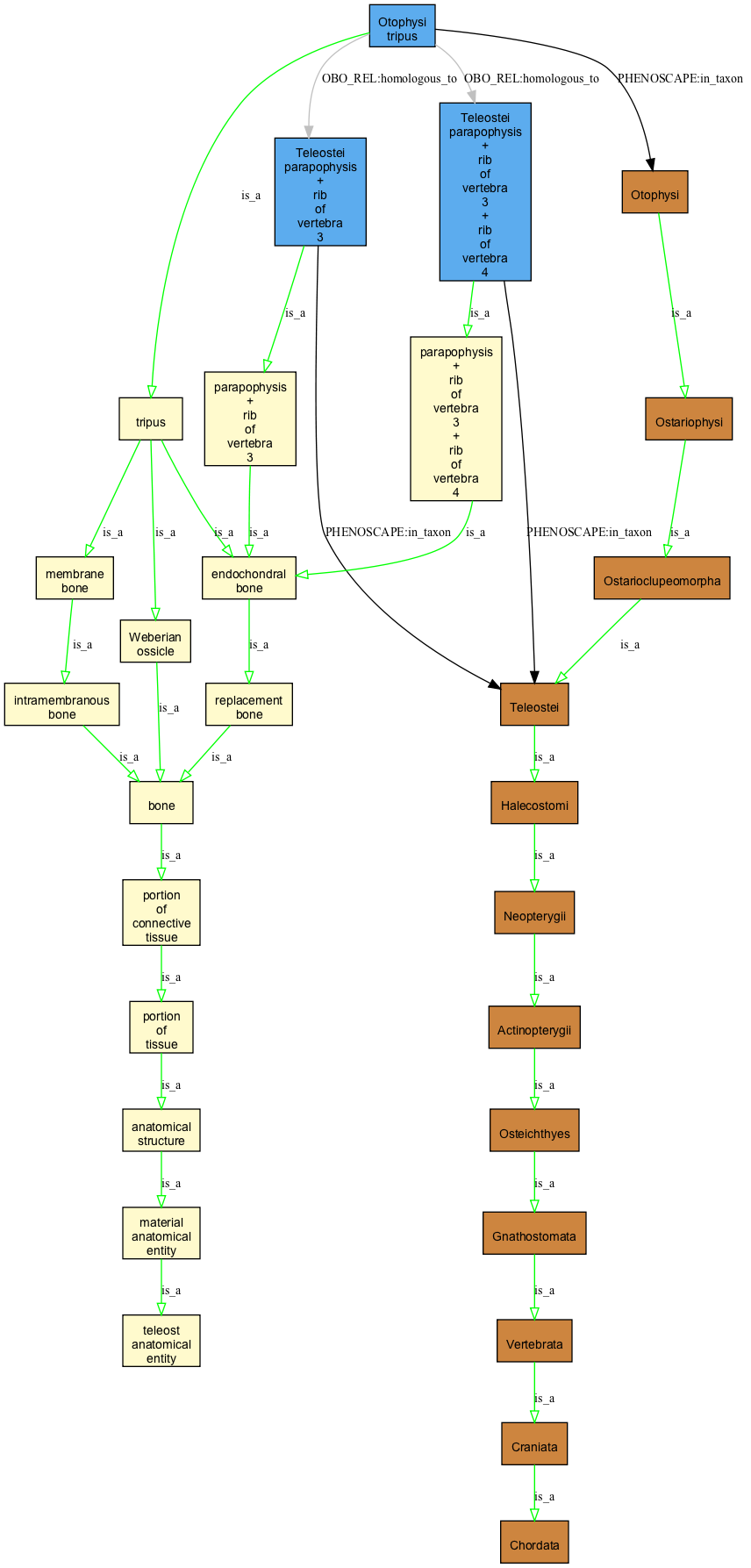
Otophysi tripus
this is annotated to be homologous to (i) the fusion of parapophysis + rib of v3 and (ii) the fusion of parapophysis + rib of v3 + rib of v4 in Teleosti. What can be said from this of the relation between (i) and (ii)