Difference between revisions of "Taxonomy ontology"
(→Resources) |
(→Alternative Designs) |
||
| (23 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
| − | The | + | This page contains accumulated design notes for taxonomy ontologies. They focus on the [[Teleost Taxonomy Ontology]] and the [[Taxonomic Rank Vocabulary]], but the most recent information for each will appear on their respective home pages. |
| + | |||
| + | The Teleost Taxonomy Ontology (TTO) is based on the Catalog of Fishes and will be updated as CoF updates appear as well as from input from area experts. The reminder of this page will discuss how the TTO is currently implemented. See the [[Taxonomic Rank Ontology]], an alternative implementation based on phylogenetic principles. | ||
| + | |||
| + | ==Organization== | ||
| + | The current organization is derived from the ontology generated from the NCBI taxonomy. There is a hierarchy of taxon names, each of which is a class. | ||
| + | |||
| + | |||
| + | |||
| + | Cyprinidae --------------------> Family | ||
| + | ^ has_rank | ||
| + | | | ||
| + | is_a | | ||
| + | | | ||
| + | Davario --------------------> Genus | ||
| + | ^ has_rank | ||
| + | | | ||
| + | is_a | | ||
| + | | | ||
| + | Davario aequipinnatus ----------------> Species | ||
| + | has_rank | ||
==Resources== | ==Resources== | ||
| Line 5: | Line 25: | ||
Here are some useful resources related to the taxonomy ontology (TTO) | Here are some useful resources related to the taxonomy ontology (TTO) | ||
| − | * [[TTO Changes|Change list (relative to November 2007 CoF update) for TTO.]] This list is current for recent changes - older changes will be added. | + | * [[TTO Changes|Change list (relative to November 2007 CoF update) for TTO.]] This list is current for recent changes - older changes will be added. |
| + | * [[Incubator | Suggested ideas for taxonomy browser and other tools]] | ||
| + | * [[Data_Jamboree_2/Notes#Taxon Concepts | Summary of a discussion of TTO issues at the second data jamboree (September 2008)]] | ||
This is a list of resources related to the development of the TTO. | This is a list of resources related to the development of the TTO. | ||
| Line 492: | Line 514: | ||
is_a: TTO:taxonomic_rank ! taxonomic_rank | is_a: TTO:taxonomic_rank ! taxonomic_rank | ||
</pre> | </pre> | ||
| + | |||
| + | =Alternative Designs= | ||
| + | Note: This section describes some design alternatives for taxonomic ontologies that refer to or include terms for ranks. | ||
| + | |||
| + | A taxonomic rank ontology allows particular taxa to be treated as historical individuals instead of as universal types. | ||
| + | |||
| + | [[Image:TaxonomicRankDiagram.png|750px|thumb|Taxonomic rank ontology and sample instance data. The ontology terms are surrounded by a grey background. The "Continuant" term would reside in an upper ontology such as [http://www.ifomis.org/bfo BFO]. The instance data constitute a particular taxonomy.]] | ||
| + | |||
| + | Previous attempts to represent taxonomy using ontology usually include taxonomic groups as classes in the ontology. Individual organisms are seen as instances of those universal types. There could be an ontology term Mammal, such that Primates and Rodents are more particular types of Mammals (''is_a'' descendants). Taxonomies are even often used as examples to help explain ontology inheritance to new users. This scheme fails to represent reality in several ways and, as this page will demonstrate, is even misleading. | ||
| + | |||
| + | ==The "traditional" view== | ||
| + | |||
| + | ===Inference with traditional taxonomy=== | ||
| + | One of the attractions of a traditional taxonomy for ontology builders that it can support the most common sort of reasoning for Description Logic ontologies. Although neither the TTO nor the NCBI taxonomy ontology attach character values to their classes, in principle such characters could be extracted from keys or from the taxonomic literature. With the addition of such properties, individuals can be classified by starting at the root of the taxonomy and traversing down the tree until either a terminal taxon (species) is reached or a node is reached where no descendant has the combination of characters displayed by the individual. For biologists, this operation is identical to use of an identification key. Such an operation is generally only defined for ''is_a'' links. | ||
| + | |||
| + | ===Problems with the "traditional" view=== | ||
| + | * The only criterion for inclusion within a taxonomic group is physical descent from a member of that group (we are assuming a taxonomy consistent with phylogeny). There are no universal properties one could ascribe to members of "Mammalia", besides things the members happen to share at this moment. "Hair" is often used an example of a Mammalian property; but of course its commonality is the result of its presence in the common ancestor of all mammals, and many mammals lack hair almost entirely. A mammal species that had no hair at all would nevertheless still be a mammal. Such reversals are common in phylogenies and can occur for both morphological properties (e.g., loss of legs in snakes) and molecular characters (e.g., inferred origin of pseudogenes). | ||
| + | * Descendant terms do not inherit (ontologically) all of the characteristics of their parents. For example, Rodentia has a different age and geographic range from Mammalia. | ||
| + | * Properties of a taxonomic group change through time: its geographic range, genetic and morphological diversity, etc. It can go extinct. These properties inhere in the group, not any one individual. | ||
| + | * Treating taxonomic groups as "classes" or "types" reinforces essentialist ideas of life forms, which evolutionary biology has completely overturned. Essentialist thought is a hindrance to understanding of the evolutionary process. | ||
| + | * Most contemporary treatments of the species concept (refs) treat them, and in many cases higher taxa, as individuals, possessing a history. | ||
| + | |||
| + | Instead of treating taxa as types in an ontology, we can treat individual taxa as instances of the type Taxon, or more specifically as instances of some taxonomic rank (see figure above). | ||
| + | |||
| + | ==Taxonomic Rank Ontology + Instance Graph of Taxonomy== | ||
| + | |||
| + | Species are increasingly seen to be better modelled as individuals. | ||
| + | |||
| + | ===Advantages=== | ||
| + | * Taxa are modeled as individuals. | ||
| + | * Taxonomic rank terms are ordered through their ''part_of'' relationships. | ||
| + | * Multiple part_of parents introduce fewer problems in inference than multiple is_a parents. This is important as more cases of reticulate phylogeny, caused by, for example, cross species hybridization or horizontal gene transfer, are discovered. | ||
| + | |||
| + | ===Inference with an ontology of taxonomic ranks + instance graph of taxa=== | ||
| + | |||
| + | ===Individuals/Instances=== | ||
| + | Implementing this approach will require [[Tool support for Instances]]. | ||
| + | |||
| + | ==Taxonomic Ranks as Metaclasses== | ||
| + | |||
| + | An approach that allows taxa to be viewed as both classes and individuals is to view rank terms, such as 'genus' as metaclasses. A metaclass is a class of classes. This would mean that a particular genus, e.g., ''Danio'' is both a subclass of Cyprinidae and an instance of genus. Metaclasses are used when assigning particular properties to a class itself, rather than its members. The property 'extinct' might be such a class-level property, as opposed to 'dead' a property that inheres in individuals. Extinct is a property we want to inhere in certain taxa only when all their individual constituents are dead. However, there are classes, such as anatomical parts, or behavior patterns that would not be subject to the possiblity of extinction. | ||
| + | |||
| + | This approach is attractive because it allows taxa to be both classes, in line with traditional taxonomies and reasoning with description logic classifiers, and individuals, with the greater fidelity to current biological thinking. | ||
| + | |||
| + | ===Disadvantages=== | ||
| + | The main disadvantage with this approach is its complexity, both for human understanding and for reasoning. | ||
| + | |||
| + | |||
| + | See also the [[Taxonomic Rank Vocabulary]] which was released in January 2010. | ||
| + | |||
| + | == Developing an Ontology for Taxonomic Ranks == | ||
| + | |||
| + | When the Teleost Taxonomy Ontology (TTO) was submitted to OBO, the suggestion was made that the terms for taxonomic ranks (e.g., Family, Genus, Species) should be broken out. At present, taxonomic ranks are included in the TTO and cross referenced to similar terms in the NCBI taxonomy ontology. Although the process of constructing an ontology of rank terms is straightforward, there are some semantic issues that need to be resolved. | ||
| + | |||
| + | The current implementation can be diagrammed as follows: | ||
| + | |||
| + | |||
| + | |||
| + | Cyprinidae --------------------> Family | ||
| + | ^ has_rank | ||
| + | | | ||
| + | is_a | | ||
| + | | | ||
| + | Devario --------------------> Genus | ||
| + | ^ has_rank | ||
| + | | | ||
| + | is_a | | ||
| + | | | ||
| + | Devario aequipinnatus ----------------> Species | ||
| + | has_rank | ||
| + | |||
| + | |||
| + | In the first draft of the ranks ontology, submitted to Chris Mungal and Michael Ashburner, the rank terms are simply subclasses of taxonomic_rank. There is no relation defined between the rank terms. The has_rank relation, as defined in the NCBO ontology is a meta_data relation (c.f. OWL annotation properties), which means it is intended to be ignored by any reasoner. | ||
| + | |||
| + | As there is clearly an ordering among the rank terms, it would be worthwhile to define an ordering relation between terms so that the term 'family' is indicated as 'larger' or 'more inclusive' than the term 'genus.' | ||
| + | |||
| + | The proposed situation is | ||
| + | |||
| + | TTO TaxonRank Ontology | ||
| + | |||
| + | Cyprinidae --------------------> Family | ||
| + | ^ has_rank ^ | ||
| + | | | | ||
| + | is_a | | "rank_order" | ||
| + | | | | ||
| + | Davario --------------------> Genus | ||
| + | ^ has_rank ^ | ||
| + | | | | ||
| + | is_a | | "rank_order" | ||
| + | | | | ||
| + | Davario aequipinnatus ----------------> Species | ||
| + | has_rank | ||
| + | |||
| + | |||
| + | There has been some question as to the nature of this ordering relation. There appear to be two points of view: | ||
| + | <ol> | ||
| + | <li>A special relation exists between taxonomic ranks. It would be transitive and antisymmetric. | ||
| + | <li>The relation is simply part_of. Part_of is transitive and antisymmetric. | ||
| + | </ol> | ||
| + | <br> | ||
| + | |||
| + | Chris Mungall's response to this question was: | ||
| + | |||
| + | 3. What about imposing an ordering among ranks and, if so, is part_of the appropriate relation? | ||
| + | |||
| + | so with any ontology you should ask "what are the instances". In this case, the instances are best considered | ||
| + | to be terms/classes/categories rather than something tangible in nature. This takes us close to weird metaclass | ||
| + | modeling territory. | ||
| + | |||
| + | but not to worry. I would say reserve part_of for "real" part_of relations, between objects and processes. I | ||
| + | would just go with a custom relation for ranks. I don't have strong opinions on what you name it - above/below? | ||
| + | more_ancestral_than? | ||
| + | |||
| + | Declare the relation transitive | ||
| + | |||
| + | There is another property of the 'rank ordering' that should be noted. The current TTO currently uses a rather limited number of taxonomic ranks. However, if this ontology is intended to be shared across OBO projects, it will need to incorporate additional taxonomic level terms. For example, the NCBI taxnomy defines over 30 rank terms. As other groups develop taxonomies they will want to add ranks that are used in their systems. Furthermore, the system of ranks is open-ended, so any particular group might need to add additional ranks in the future. However, any one taxonomy will only use a subset of these terms, which means that in most cases, the is_a relation between two taxa will frequently correspond to more than one link up the rank taxonomy. Another case where this situation comes up, even for a single taxonomy, is an incertae sedis taxa. Thus a definition for the ordering relation needs to work without depending on having every step in the rank chain correspond to a taxonomic term in each tip to root path in the tree. | ||
| + | |||
| + | |||
| + | Resolving this issue may depend on the resolution of a second, closely related issue that may have to be reopened for discussion. | ||
| + | |||
| + | == Modeling Taxa == | ||
| + | |||
| + | Following the example of the OBO translation of the NCBI taxonomy, the TTO models taxa as a hierarchy of classes, defined by an is_a relation based on set theory. This means that classes, hence taxa terms are represented as sets. The taxon as set model extends all the way down to and including the species level. | ||
| + | |||
| + | However, an increasingly influential view among philosophers of biology is that species should be not be seen as either classes (or natural kinds), but as evolutionary units, and hence as individuals (e.g., Hull 1974, Ghiselin 1978). In this view, species are not sets of individual organisms, rather there is a part_of relation between organisms and their species. The particular part_of relation is commonly portrayed as species (and other clades) being composed of lineages and lineages consisting of related individual organisms. | ||
| + | |||
| + | This view can be extended to consider clades as individuals, since they, like species are comprised of lineages. Individuals comprised of lineages might be modeled as instances of the class 'portion of clade', which might include species (the class of all individual species) as a particular subclass because individual species are evolutionary units, unlike larger clades. | ||
| + | |||
| + | Modeled this way, the TTO and taxon rank ontology might look as follows: | ||
| + | |||
| + | TTO TaxonRank Ontology | ||
| + | |||
| + | |||
| + | is_a | ||
| + | "portion of clade" <-------------- Cyprinidae --------------------> Family | ||
| + | ^ has_rank ^ | ||
| + | | | | ||
| + | part_of | | "rank_order" | ||
| + | is_a | | | ||
| + | "portion of clade" <----------- Davario --------------------> Genus | ||
| + | ^ has_rank ^ | ||
| + | | | | ||
| + | part_of | | "rank_order" | ||
| + | is_a | | | ||
| + | "portion of clade" <------- Davario aequipinnatus ----------------> Species | ||
| + | has_rank | ||
| + | |||
| + | |||
| + | Or even: | ||
| + | |||
| + | |||
| + | TTO TaxonRank Ontology | ||
| + | |||
| + | |||
| + | is_a is_a | ||
| + | Cyprinidae --------------------> Family ------------------> "portion of clade" | ||
| + | ^ ^ | ||
| + | | | | ||
| + | | part_of | "rank_order" | ||
| + | | is_a | | ||
| + | Davario --------------------> Genus ------------------> "portion of clade" | ||
| + | ^ ^ is_a | ||
| + | | | | ||
| + | | part_of | "rank_order" | ||
| + | | is_a | | ||
| + | Davario aequipinnatus ------------> Species -----------------> "portion of clade" | ||
| + | |||
| + | |||
| + | This approach also provides a natural bridge to the use of Phylogenetic definitions, which more or less restrict taxonomic labels to monophyletic clades. A more detailed description of this approach can be found [[Taxonomic Rank Ontology|here]]. | ||
| + | |||
| + | The major difficulties with such a model are potential conflicts with the OBO ontologies focus on classes, rather than individuals. | ||
| + | |||
| + | =Other Notes= | ||
| + | ;1. Taxonomic ranks - keep in TTO or make into separate ontology? | ||
| + | - This issue has been resolved see [[Taxonomic Rank Vocabulary]]. | ||
| + | |||
| + | ;2. Representing unknown/unnamed species in ontology | ||
| + | :a. Examples: | ||
| + | :i. Chela sp. | ||
| + | :ii. Esomus sp. | ||
| + | : iii. Danio aff. dangila | ||
| + | :b. Solution? Instead of assigning a rank, create new entities for these, with appropriate genus as the parent and reference publication information, possibly in the name, e.g. ‘Chela sp. (Fang 2003)’ | ||
| + | |||
| + | - We decided to follow solution given in 2b. We will call these species whatever the author calls them, i.e. if Fang 2003 calls a species ‘Danio aff. dangila’, we will request the name ‘Danio aff. dangila (Fang 2003)’ from Peter. | ||
| + | |||
| + | - We will make requests for new species names through the TTO Tracker (vs. email to Peter. | ||
| + | |||
| + | - If there are multiple entries for an unknown species in the materials list (e.g., ‘Chela sp.’ is listed multiple times with different voucher numbers), we will create only one entity for the unknown species. If in the future these multiple vouchers are identified as different species, then we will update our annotations to reflect the new species designation. | ||
| + | |||
| + | - All these names will be assigned ‘high’ ids, pending their incorporation into CoF or demotion to synonyms as appropriate in the future. | ||
| + | |||
| + | |||
| + | ;3. Representing incertae sedis in ontology | ||
| + | |||
| + | - We will represent incertae sedis as currently done (point species directly to appropriate level in ontology) | ||
| + | |||
| + | - We discussed creating duplicate taxonomic category for incertae sedis (for example, ‘Cyprinidae incertae sedis’) but this implies that all species within this category are a family and we want to avoid that | ||
| + | |||
| + | - We also discussed creating new taxonomic ranks for incertae sedis (e.g. Family incertae sedis, Order incertae sedis) to which we would annotate incertae sedis taxa (vs. assigning e.g. Order or Family rank); creates categories that are not ontologically sound, so no. | ||
| + | |||
| + | - Issue with representing uncertainly monophyletic genera – we think we can deal with this by giving users the options to view trees with alternative taxonomy for these species | ||
| + | |||
| + | - Tree views will solve many incertae sedis issues. | ||
| + | |||
| + | - I’m not sure how this will interact with groups where intermediate levels are added – lots of families might be incertae sedis at the level of tribe for example – (Peter). | ||
| + | |||
| + | |||
| + | ;4. Incomplete tracking of synonyms from CoF to TTO | ||
| + | |||
| + | - Synonym extraction has been implemented in the [[TTOUpdate tool]]. There is still room for improvement in the extraction algorithm. | ||
| + | |||
| + | |||
| + | ;5. Error reports to Peter – continue to send by email? Thoughts to setting up a tracker? | ||
| + | |||
| + | - There is a tracker for requests and error reports within the [https://sourceforge.net/tracker/?atid=1046550&group_id=76834&func=browse OBO sourceforge site]. | ||
| + | |||
| + | [[Category:Ontology]] | ||
| + | [[Category:Taxonomy]] | ||
Latest revision as of 17:01, 31 August 2010
This page contains accumulated design notes for taxonomy ontologies. They focus on the Teleost Taxonomy Ontology and the Taxonomic Rank Vocabulary, but the most recent information for each will appear on their respective home pages.
The Teleost Taxonomy Ontology (TTO) is based on the Catalog of Fishes and will be updated as CoF updates appear as well as from input from area experts. The reminder of this page will discuss how the TTO is currently implemented. See the Taxonomic Rank Ontology, an alternative implementation based on phylogenetic principles.
Contents
- 1 Organization
- 2 Resources
- 3 Taxonomy ontology structure
- 4 Taxonomy ontology content from Catalog of Fishes
- 5 Alternative Designs
- 6 Other Notes
Organization
The current organization is derived from the ontology generated from the NCBI taxonomy. There is a hierarchy of taxon names, each of which is a class.
Cyprinidae --------------------> Family
^ has_rank
|
is_a |
|
Davario --------------------> Genus
^ has_rank
|
is_a |
|
Davario aequipinnatus ----------------> Species
has_rank
Resources
Here are some useful resources related to the taxonomy ontology (TTO)
- Change list (relative to November 2007 CoF update) for TTO. This list is current for recent changes - older changes will be added.
- Suggested ideas for taxonomy browser and other tools
- Summary of a discussion of TTO issues at the second data jamboree (September 2008)
This is a list of resources related to the development of the TTO.
- Chris Mungall's NCBI Taxonomy in OBO format - this file is huge!
- Catalog of Fishes database schema documentation. - possibly somewhat out of date
- OBO Flat File Format Specification
- OBO Identifier Lifecycle
Taxonomy ontology structure
There is now a page devoted to discussion of Taxonomic Ranks.
The NCBI taxonomy ontology has some special terms and relations which are probably relevant to the fish taxonomy ontology. One of these is the "has_rank" typedef, which is used as a property value on the taxon terms:
[Typedef] id: has_rank name: has_rank def: "A metadata relation between a class and its taxonomic rank (eg species, family)" [] comment: This is an abstract class for use with the NCBI taxonomy to name the depth of the node within the tree. The link between the node term and the rank is only visible if you are using an obo 1.3 aware browser/editor; otherwise this can be ignored is_metadata_tag: true
Some special terms are included which represent taxonomic ranks. They descend from the term "taxonomic_rank":
[Term] id: NCBITaxon:taxonomic_rank name: taxonomic_rank def: "A level of depth within a species taxonomic tree" [] comment: This is an abstract class for use with the NCBI taxonomy to name the depth of the node within the tree. The link between the node term and the rank is only visible if you are using an obo 1.3 aware browser/editor; otherwise this can be ignored [Term] id: NCBITaxon:superkingdom name: superkingdom is_a: NCBITaxon:taxonomic_rank [Term] id: NCBITaxon:genus name: genus is_a: NCBITaxon:taxonomic_rank [Term] id: NCBITaxon:species name: species is_a: NCBITaxon:taxonomic_rank etc....
Here are three actual taxon terms. The term for the species descends via is_a from the term for the genus, and likewise for the genus and family. The rank is specified in the property_value. Other names are placed in the synonym fields:
[Term] id: NCBITaxon:7996 name: Ictaluridae is_a: NCBITaxon:7995 synonym: "North American freshwater catfishes" EXACT common_name [] synonym: "bullhead catfishes" EXACT genbank_common_name [] property_value: has_rank NCBITaxon:family xref: GC_ID:1 [Term] id: NCBITaxon:7997 name: Ictalurus is_a: NCBITaxon:7996 property_value: has_rank NCBITaxon:genus xref: GC_ID:1 [Term] id: NCBITaxon:7998 name: Ictalurus punctatus is_a: NCBITaxon:7997 synonym: "channel catfish" EXACT genbank_common_name [] property_value: has_rank NCBITaxon:species xref: GC_ID:1
Other uses for the synonym field can be seen in the OBO file, such as for obsolete names. We may want to also provide a definition from the original species description.
Taxonomy ontology content from Catalog of Fishes
Which parts of the CoF schema should be included in the taxonomy ontology? Also, is there information not in the Catalog of Fishes that should be included in the taxonomy ontology?
There is now a rough draft Taxonomy ontology directly from (somewhat dated) dump files from the Catalog of Fishes.
A draft version with synonyms is here. The actual top of the tree is still Craniata, other species
appearing at the root represent problems - usually related to synonyms that we need to resolve. Also note that I have tagged all
synonyms as "RELATED", the weakest category of synonymy. Given the complexities of taxonomic synonymy, this seems to be the appropriate
place to start. With the most recent version, most synonyms also include database references (dbxrefs) back to the original genus and species id's from the CoF tables.
Here is a preliminary taxonomy ontology, using data from a somewhat dated CoF dump. The taxa represented here are mentioned in
Fink, S.V., and W. L. Fink. 1981. Interrelationships of the ostariophysan fishes (Teleostei). Zoological Journal of the Linnean
Society 72: 297–353.
Craniata is included as a root for the entire taxonomy, even though it does not correspond to any taxonomic level defined in the ontology.
Note also that the term ids are not generated by OBO edit, we are basing them on catalog numbers from CoF.
Craniata: 1 Our root
Class: 10 + the class number from CoF (are these subject to splits?)
Order: 1000 + 10 * the order number from CoF - these numbers are sometimes formatted XX.X (to indicate splits?)
Family: 10000 + 10 * the family number from CoF - these numbers are sometimes formatted XX.X (to indicate splits?)
Genus: 100000 + the CAS_GEN value from CoF
Species: 1000000 + the CAS_SPC value from CoF
Unnamed entities: 10000000+ (e.g., sp., spp., etc.)
We should probably discuss whether this id scheme is really what we want to do - it means we can't add taxa from OBOEdit, though that isn't necessarily a problem.
format-version: 1.2 date: 02:10:2007 15:27 saved-by: peter auto-generated-by: OBO-Edit 1.101 default-namespace: teleost-taxonomy [Term] id: TTO: 1029899 name: Prochilodus vimboides is_a: TTO:101208 ! Prochilodus property_value: has_rank TTO:species [Term] id: TTO:1 name: Craniata [Term] id: TTO:10000001 name: Eigenmannia sp. is_a: TTO:104678 ! Eigenmannia property_value: has_rank TTO:Unknown_species [Term] id: TTO:10000002 name: Sternopygus sp. is_a: TTO:102060 ! Sternopygus property_value: has_rank TTO:Unknown_species [Term] id: TTO:1003114 name: Notemigonus crysoleucus is_a: TTO:100967 ! Notemigonus property_value: has_rank TTO:species [Term] id: TTO:1003239 name: Opsariichthys uncirostris is_a: TTO:103159 ! Opsariichthys property_value: has_rank TTO:species [Term] id: TTO:1004143 name: Chalceus macrolepidotus is_a: TTO:100825 ! Chalceus property_value: has_rank TTO:species [Term] id: TTO:1004205 name: Hepsetus odoe is_a: TTO:109162 ! Hepsetus property_value: has_rank TTO:species [Term] id: TTO:1004213 name: Xenocharax spilurus is_a: TTO:103438 ! Xenocharax property_value: has_rank TTO:species [Term] id: TTO:1004343 name: Rhoadsia altipinna is_a: TTO:105589 ! Rhoadsia property_value: has_rank TTO:species [Term] id: TTO:100558 name: Chanos is_a: TTO:10720 ! Chanidae property_value: has_rank TTO:genus [Term] id: TTO:1006103 name: Rhabdolichops troscheli is_a: TTO:107098 ! Rhabdolichops property_value: has_rank TTO:species [Term] id: TTO:100825 name: Chalceus is_a: TTO:10910 ! Characidae property_value: has_rank TTO:genus [Term] id: TTO:100967 name: Notemigonus is_a: TTO:10760 ! Cyprinidae property_value: has_rank TTO:genus [Term] id: TTO:101208 name: Prochilodus is_a: TTO:10855 ! Prochilodontidae property_value: has_rank TTO:genus [Term] id: TTO:101842 name: Brycon is_a: TTO:10910 ! Characidae property_value: has_rank TTO:genus [Term] id: TTO:102060 name: Sternopygus is_a: TTO:11231 ! Sternopygidae property_value: has_rank TTO:genus [Term] id: TTO:1027661 name: Chanos chanos is_a: TTO:100558 ! Chanos property_value: has_rank TTO:species [Term] id: TTO:1028504 name: Bryconamericus brevirostris is_a: TTO:105385 ! Bryconamericus property_value: has_rank TTO:species [Term] id: TTO:102915 name: Diplomystes is_a: TTO:10920 ! Diplomystidae property_value: has_rank TTO:genus [Term] id: TTO:1029536 name: Brycon dentex is_a: TTO:101842 ! Brycon property_value: has_rank TTO:species [Term] id: TTO:1030110 name: Platyurosternarchus macrostomus synonym: "Sternarchus macrostoma" EXACT [] is_a: TTO:110391 ! Platyurosternarchus property_value: has_rank TTO:species [Term] id: TTO:103159 name: Opsariichthys is_a: TTO:10760 ! Cyprinidae property_value: has_rank TTO:genus [Term] id: TTO:103339 name: Auchenoglanis is_a: TTO:10940 ! Bagridae property_value: has_rank TTO:genus [Term] id: TTO:103438 name: Xenocharax is_a: TTO:10820 ! Citharinidae property_value: has_rank TTO:genus [Term] id: TTO:1046341 name: Diplomystes chilensis synonym: "Diplomystes papillosus" EXACT [] is_a: TTO:102915 ! Diplomystes property_value: has_rank TTO:species [Term] id: TTO:104678 name: Eigenmannia is_a: TTO:11231 ! Sternopygidae property_value: has_rank TTO:genus [Term] id: TTO:105055 name: Zacco is_a: TTO:10760 ! Cyprinidae property_value: has_rank TTO:genus [Term] id: TTO:1051779 name: Zacco temminckii is_a: TTO:105055 ! Zacco property_value: has_rank TTO:species [Term] id: TTO:105268 name: Sternarchorhamphus is_a: TTO:11232 ! Apteronotidae property_value: has_rank TTO:genus [Term] id: TTO:105385 name: Bryconamericus is_a: TTO:10910 ! Characidae property_value: has_rank TTO:genus [Term] id: TTO:105589 name: Rhoadsia is_a: TTO:10910 ! Characidae property_value: has_rank TTO:genus [Term] id: TTO:1056213 name: Auchenoglanis monkei synonym: "Parauchenoglanis guttatus" EXACT [] is_a: TTO:103339 ! Auchenoglanis property_value: has_rank TTO:species [Term] id: TTO:1062171 name: Sternopygus macrurus is_a: TTO:102060 ! Sternopygus property_value: has_rank TTO:species [Term] id: TTO:107098 name: Rhabdolichops is_a: TTO:11231 ! Sternopygidae property_value: has_rank TTO:genus [Term] id: TTO:10720 name: Chanidae is_a: TTO:1350 ! Gonorynchiformes property_value: has_rank TTO:family [Term] id: TTO:10760 name: Cyprinidae is_a: TTO:1360 ! Cypriniformes property_value: has_rank TTO:family [Term] id: TTO:10820 name: Citharinidae is_a: TTO:1370 ! Characiformes property_value: has_rank TTO:family [Term] id: TTO:10827 name: Hepsetidae is_a: TTO:1370 ! Characiformes property_value: has_rank TTO:family [Term] id: TTO:10855 name: Prochilodontidae is_a: TTO:1370 ! Characiformes property_value: has_rank TTO:family [Term] id: TTO:10910 name: Characidae is_a: TTO:1370 ! Characiformes property_value: has_rank TTO:family [Term] id: TTO:109162 name: Hepsetus is_a: TTO:10827 ! Hepsetidae property_value: has_rank TTO:genus [Term] id: TTO:10920 name: Diplomystidae is_a: TTO:1380 ! Siluriformes property_value: has_rank TTO:family [Term] id: TTO:10940 name: Bagridae is_a: TTO:1380 ! Siluriformes property_value: has_rank TTO:family [Term] id: TTO:110391 name: Platyurosternarchus is_a: TTO:11232 ! Apteronotidae property_value: has_rank TTO:genus [Term] id: TTO:11231 name: Sternopygidae is_a: TTO:1390 ! Gymnotiformes property_value: has_rank TTO:family [Term] id: TTO:11232 name: Apteronotidae is_a: TTO:1390 ! Gymnotiformes property_value: has_rank TTO:family [Term] id: TTO:1350 name: Gonorynchiformes is_a: TTO:18 ! Actinopterygii property_value: has_rank TTO:order [Term] id: TTO:1360 name: Cypriniformes is_a: TTO:18 ! Actinopterygii property_value: has_rank TTO:order [Term] id: TTO:1370 name: Characiformes is_a: TTO:18 ! Actinopterygii property_value: has_rank TTO:order [Term] id: TTO:1380 name: Siluriformes is_a: TTO:18 ! Actinopterygii property_value: has_rank TTO:order [Term] id: TTO:1390 name: Gymnotiformes is_a: TTO:18 ! Actinopterygii property_value: has_rank TTO:order [Term] id: TTO:18 name: Actinopterygii is_a: TTO:1 ! Craniata property_value: has_rank TTO:class [Term] id: TTO:class name: class xref: NCBITaxon:class is_a: TTO:taxonomic_rank ! taxonomic_rank [Term] id: TTO:family name: family xref: NCBITaxon:family is_a: TTO:taxonomic_rank ! taxonomic_rank [Term] id: TTO:genus name: genus xref: NCBITaxon:genus is_a: TTO:taxonomic_rank ! taxonomic_rank [Term] id: TTO:order name: order xref: NCBITaxon:order is_a: TTO:taxonomic_rank ! taxonomic_rank [Term] id: TTO:species name: species xref: NCBITaxon:species is_a: TTO:taxonomic_rank ! taxonomic_rank [Term] id: TTO:taxonomic_rank name: taxonomic_rank xref: NCBITaxon:taxonomic_rank [Term] id: TTO:Unknown_species name: Unknown species is_a: TTO:taxonomic_rank ! taxonomic_rank [Term] id: TTO:Unknown_species_group name: Unknown species group is_a: TTO:taxonomic_rank ! taxonomic_rank [Term] id: TTO:Unnamed_species name: Unnamed species is_a: TTO:taxonomic_rank ! taxonomic_rank
Alternative Designs
Note: This section describes some design alternatives for taxonomic ontologies that refer to or include terms for ranks.
A taxonomic rank ontology allows particular taxa to be treated as historical individuals instead of as universal types.
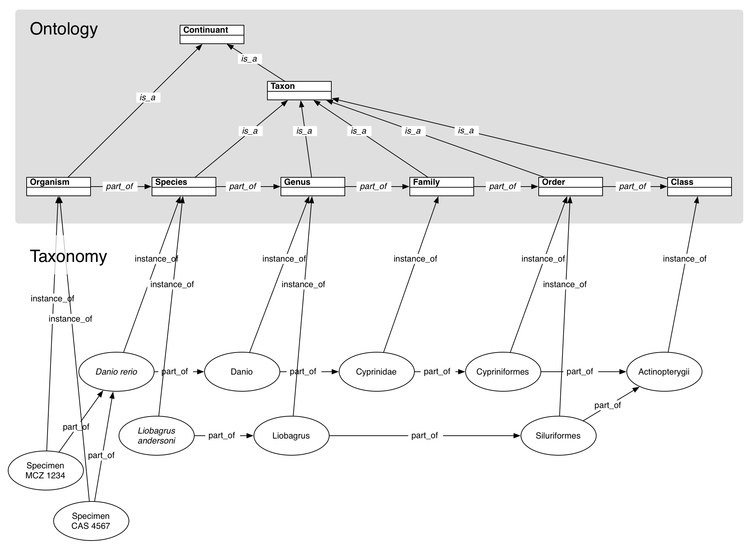
Previous attempts to represent taxonomy using ontology usually include taxonomic groups as classes in the ontology. Individual organisms are seen as instances of those universal types. There could be an ontology term Mammal, such that Primates and Rodents are more particular types of Mammals (is_a descendants). Taxonomies are even often used as examples to help explain ontology inheritance to new users. This scheme fails to represent reality in several ways and, as this page will demonstrate, is even misleading.
The "traditional" view
Inference with traditional taxonomy
One of the attractions of a traditional taxonomy for ontology builders that it can support the most common sort of reasoning for Description Logic ontologies. Although neither the TTO nor the NCBI taxonomy ontology attach character values to their classes, in principle such characters could be extracted from keys or from the taxonomic literature. With the addition of such properties, individuals can be classified by starting at the root of the taxonomy and traversing down the tree until either a terminal taxon (species) is reached or a node is reached where no descendant has the combination of characters displayed by the individual. For biologists, this operation is identical to use of an identification key. Such an operation is generally only defined for is_a links.
Problems with the "traditional" view
- The only criterion for inclusion within a taxonomic group is physical descent from a member of that group (we are assuming a taxonomy consistent with phylogeny). There are no universal properties one could ascribe to members of "Mammalia", besides things the members happen to share at this moment. "Hair" is often used an example of a Mammalian property; but of course its commonality is the result of its presence in the common ancestor of all mammals, and many mammals lack hair almost entirely. A mammal species that had no hair at all would nevertheless still be a mammal. Such reversals are common in phylogenies and can occur for both morphological properties (e.g., loss of legs in snakes) and molecular characters (e.g., inferred origin of pseudogenes).
- Descendant terms do not inherit (ontologically) all of the characteristics of their parents. For example, Rodentia has a different age and geographic range from Mammalia.
- Properties of a taxonomic group change through time: its geographic range, genetic and morphological diversity, etc. It can go extinct. These properties inhere in the group, not any one individual.
- Treating taxonomic groups as "classes" or "types" reinforces essentialist ideas of life forms, which evolutionary biology has completely overturned. Essentialist thought is a hindrance to understanding of the evolutionary process.
- Most contemporary treatments of the species concept (refs) treat them, and in many cases higher taxa, as individuals, possessing a history.
Instead of treating taxa as types in an ontology, we can treat individual taxa as instances of the type Taxon, or more specifically as instances of some taxonomic rank (see figure above).
Taxonomic Rank Ontology + Instance Graph of Taxonomy
Species are increasingly seen to be better modelled as individuals.
Advantages
- Taxa are modeled as individuals.
- Taxonomic rank terms are ordered through their part_of relationships.
- Multiple part_of parents introduce fewer problems in inference than multiple is_a parents. This is important as more cases of reticulate phylogeny, caused by, for example, cross species hybridization or horizontal gene transfer, are discovered.
Inference with an ontology of taxonomic ranks + instance graph of taxa
Individuals/Instances
Implementing this approach will require Tool support for Instances.
Taxonomic Ranks as Metaclasses
An approach that allows taxa to be viewed as both classes and individuals is to view rank terms, such as 'genus' as metaclasses. A metaclass is a class of classes. This would mean that a particular genus, e.g., Danio is both a subclass of Cyprinidae and an instance of genus. Metaclasses are used when assigning particular properties to a class itself, rather than its members. The property 'extinct' might be such a class-level property, as opposed to 'dead' a property that inheres in individuals. Extinct is a property we want to inhere in certain taxa only when all their individual constituents are dead. However, there are classes, such as anatomical parts, or behavior patterns that would not be subject to the possiblity of extinction.
This approach is attractive because it allows taxa to be both classes, in line with traditional taxonomies and reasoning with description logic classifiers, and individuals, with the greater fidelity to current biological thinking.
Disadvantages
The main disadvantage with this approach is its complexity, both for human understanding and for reasoning.
See also the Taxonomic Rank Vocabulary which was released in January 2010.
Developing an Ontology for Taxonomic Ranks
When the Teleost Taxonomy Ontology (TTO) was submitted to OBO, the suggestion was made that the terms for taxonomic ranks (e.g., Family, Genus, Species) should be broken out. At present, taxonomic ranks are included in the TTO and cross referenced to similar terms in the NCBI taxonomy ontology. Although the process of constructing an ontology of rank terms is straightforward, there are some semantic issues that need to be resolved.
The current implementation can be diagrammed as follows:
Cyprinidae --------------------> Family
^ has_rank
|
is_a |
|
Devario --------------------> Genus
^ has_rank
|
is_a |
|
Devario aequipinnatus ----------------> Species
has_rank
In the first draft of the ranks ontology, submitted to Chris Mungal and Michael Ashburner, the rank terms are simply subclasses of taxonomic_rank. There is no relation defined between the rank terms. The has_rank relation, as defined in the NCBO ontology is a meta_data relation (c.f. OWL annotation properties), which means it is intended to be ignored by any reasoner.
As there is clearly an ordering among the rank terms, it would be worthwhile to define an ordering relation between terms so that the term 'family' is indicated as 'larger' or 'more inclusive' than the term 'genus.'
The proposed situation is
TTO TaxonRank Ontology
Cyprinidae --------------------> Family
^ has_rank ^
| |
is_a | | "rank_order"
| |
Davario --------------------> Genus
^ has_rank ^
| |
is_a | | "rank_order"
| |
Davario aequipinnatus ----------------> Species
has_rank
There has been some question as to the nature of this ordering relation. There appear to be two points of view:
- A special relation exists between taxonomic ranks. It would be transitive and antisymmetric.
- The relation is simply part_of. Part_of is transitive and antisymmetric.
Chris Mungall's response to this question was:
3. What about imposing an ordering among ranks and, if so, is part_of the appropriate relation?
so with any ontology you should ask "what are the instances". In this case, the instances are best considered to be terms/classes/categories rather than something tangible in nature. This takes us close to weird metaclass modeling territory.
but not to worry. I would say reserve part_of for "real" part_of relations, between objects and processes. I would just go with a custom relation for ranks. I don't have strong opinions on what you name it - above/below? more_ancestral_than?
Declare the relation transitive
There is another property of the 'rank ordering' that should be noted. The current TTO currently uses a rather limited number of taxonomic ranks. However, if this ontology is intended to be shared across OBO projects, it will need to incorporate additional taxonomic level terms. For example, the NCBI taxnomy defines over 30 rank terms. As other groups develop taxonomies they will want to add ranks that are used in their systems. Furthermore, the system of ranks is open-ended, so any particular group might need to add additional ranks in the future. However, any one taxonomy will only use a subset of these terms, which means that in most cases, the is_a relation between two taxa will frequently correspond to more than one link up the rank taxonomy. Another case where this situation comes up, even for a single taxonomy, is an incertae sedis taxa. Thus a definition for the ordering relation needs to work without depending on having every step in the rank chain correspond to a taxonomic term in each tip to root path in the tree.
Resolving this issue may depend on the resolution of a second, closely related issue that may have to be reopened for discussion.
Modeling Taxa
Following the example of the OBO translation of the NCBI taxonomy, the TTO models taxa as a hierarchy of classes, defined by an is_a relation based on set theory. This means that classes, hence taxa terms are represented as sets. The taxon as set model extends all the way down to and including the species level.
However, an increasingly influential view among philosophers of biology is that species should be not be seen as either classes (or natural kinds), but as evolutionary units, and hence as individuals (e.g., Hull 1974, Ghiselin 1978). In this view, species are not sets of individual organisms, rather there is a part_of relation between organisms and their species. The particular part_of relation is commonly portrayed as species (and other clades) being composed of lineages and lineages consisting of related individual organisms.
This view can be extended to consider clades as individuals, since they, like species are comprised of lineages. Individuals comprised of lineages might be modeled as instances of the class 'portion of clade', which might include species (the class of all individual species) as a particular subclass because individual species are evolutionary units, unlike larger clades.
Modeled this way, the TTO and taxon rank ontology might look as follows:
TTO TaxonRank Ontology
is_a
"portion of clade" <-------------- Cyprinidae --------------------> Family
^ has_rank ^
| |
part_of | | "rank_order"
is_a | |
"portion of clade" <----------- Davario --------------------> Genus
^ has_rank ^
| |
part_of | | "rank_order"
is_a | |
"portion of clade" <------- Davario aequipinnatus ----------------> Species
has_rank
Or even:
TTO TaxonRank Ontology
is_a is_a
Cyprinidae --------------------> Family ------------------> "portion of clade"
^ ^
| |
| part_of | "rank_order"
| is_a |
Davario --------------------> Genus ------------------> "portion of clade"
^ ^ is_a
| |
| part_of | "rank_order"
| is_a |
Davario aequipinnatus ------------> Species -----------------> "portion of clade"
This approach also provides a natural bridge to the use of Phylogenetic definitions, which more or less restrict taxonomic labels to monophyletic clades. A more detailed description of this approach can be found here.
The major difficulties with such a model are potential conflicts with the OBO ontologies focus on classes, rather than individuals.
Other Notes
- 1. Taxonomic ranks - keep in TTO or make into separate ontology?
- This issue has been resolved see Taxonomic Rank Vocabulary.
- 2. Representing unknown/unnamed species in ontology
- a. Examples:
- i. Chela sp.
- ii. Esomus sp.
- iii. Danio aff. dangila
- b. Solution? Instead of assigning a rank, create new entities for these, with appropriate genus as the parent and reference publication information, possibly in the name, e.g. ‘Chela sp. (Fang 2003)’
- We decided to follow solution given in 2b. We will call these species whatever the author calls them, i.e. if Fang 2003 calls a species ‘Danio aff. dangila’, we will request the name ‘Danio aff. dangila (Fang 2003)’ from Peter.
- We will make requests for new species names through the TTO Tracker (vs. email to Peter.
- If there are multiple entries for an unknown species in the materials list (e.g., ‘Chela sp.’ is listed multiple times with different voucher numbers), we will create only one entity for the unknown species. If in the future these multiple vouchers are identified as different species, then we will update our annotations to reflect the new species designation.
- All these names will be assigned ‘high’ ids, pending their incorporation into CoF or demotion to synonyms as appropriate in the future.
- 3. Representing incertae sedis in ontology
- We will represent incertae sedis as currently done (point species directly to appropriate level in ontology)
- We discussed creating duplicate taxonomic category for incertae sedis (for example, ‘Cyprinidae incertae sedis’) but this implies that all species within this category are a family and we want to avoid that
- We also discussed creating new taxonomic ranks for incertae sedis (e.g. Family incertae sedis, Order incertae sedis) to which we would annotate incertae sedis taxa (vs. assigning e.g. Order or Family rank); creates categories that are not ontologically sound, so no.
- Issue with representing uncertainly monophyletic genera – we think we can deal with this by giving users the options to view trees with alternative taxonomy for these species
- Tree views will solve many incertae sedis issues.
- I’m not sure how this will interact with groups where intermediate levels are added – lots of families might be incertae sedis at the level of tribe for example – (Peter).
- 4. Incomplete tracking of synonyms from CoF to TTO
- Synonym extraction has been implemented in the TTOUpdate tool. There is still room for improvement in the extraction algorithm.
- 5. Error reports to Peter – continue to send by email? Thoughts to setting up a tracker?
- There is a tracker for requests and error reports within the OBO sourceforge site.